
扩展功能
文章信息
- 孟娇, 陈晶瑜
- MENG Jiao, CHEN Jingyu
- Rcs双组分调节系统对细菌环境应答的分子调控研究进展
- Role of Rcs two-component regulatory system in environmental responses of bacteria: a review
- 微生物学通报, 2021, 48(1): 210-222
- Microbiology China, 2021, 48(1): 210-222
- DOI: 10.13344/j.microbiol.china.200477
-
文章历史
- 收稿日期: 2020-05-15
- 接受日期: 2020-07-13
- 网络首发日期: 2020-09-22
双组分信号转导系统是细菌感应外界信号、广谱且精细调控多种生命活动的基因表达调控系统[1-2]。根据磷酸传递步骤的不同,双组分信号转导系统可分为典型和非典型系统。不论是哪一类系统,双组分信号转导系统均由组氨酸蛋白激酶(Histidine Kinase,HK)和反应调节蛋白(Response Regulator Protein,RR)这2种成分组成[2]。典型双组分系统的磷酸传递过程是从HK到RR仅需一步就可完成,而非典型双组分系统需要多步磷酸传递过程,磷酸接力传递过程需要在多个His和Asp残基之间交替进行[2]。Rcs系统最初在1985年被确定为大肠杆菌(Escherichia coli)荚膜多糖可拉酸(Colanic Acid)生物合成的正调控因子[3]。随后的研究表明,Rcs系统是存在于革兰氏阴性细菌肠杆菌科成员中的非典型双组分调节系统(Two-Component Regulatory System,TCS)[4]。在渗透压、温度、pH、金属离子、溶菌酶、细胞缺陷等多种环境信号分子的作用下,通过作用于鞭毛、生物被膜、毒力相关基因等多个靶标,调控运动、生物被膜形成、毒力等多种生命活动[5]。近年来对细菌Rcs系统环境应答机制的探索成为一个新的研究热点[6-11]。本文对Rcs系统在细菌感知环境信号和调节基因表达方面进行了归纳、总结,以期增进对细菌Rcs系统的了解,为进一步的研究提供依据,也可为细菌的控制、预防及治疗方案的开发提供参考。
1 Rcs系统的概述Rcs系统由跨膜感应激酶RcsC、跨膜蛋白RcsD和响应调节剂RcsB这3个核心蛋白组成[1, 4, 12]。RcsC传感器同时具有激酶和磷酸酶活性,与响应调节剂RcsB共同代表了细菌TCS的经典成员,而膜结合传感器RcsD则缺乏激酶活性[4, 12]。在没有任何环境信号的作用下,RcsC与RcsD一起担任磷酸酶的作用,确保细胞中磷酸化的RcsB (RcsB-P)保持在低水平[12]。RcsA是一种辅助蛋白,可在其标记为RcsAB的位点与RcsB结合,从而调控基因的表达[13]。除了RcsA之外,还发现了BglJ、GadE、MatA (EcpR)、DctR、TviA、RflM和RmpA等多种辅助蛋白[4]。外膜脂蛋白RcsF将由膜应激或其他元件诱导的信号传递到RcsC,从而激活Rcs系统[14]。
Rcs系统磷酸信号的传递过程一般遵循His-Asp-His-Asp路径(图 1)。以E.coli为例,外膜脂蛋白RcsF将由膜应激或其他元件诱导的信号传递给RcsC,在ATP的参与下,RcsC组氨酸激酶功能域HK保守的His479发生自身磷酸化,随后被自身磷酸接受功能域(Phosphoryl Receiver,PR)上的天冬氨酸残基Asp875接受,磷酸信号传递至RcsD磷酸信号传递功能域(Histidine-Containing Phosphotransmitter,HPT)上的组氨酸残基His842,最终传递至转录调控因子RcsB的天冬氨酸残基Asp56[1, 4, 12]。此外,在缺乏外界刺激或对某些代谢应激做出反应时,RcsB也可被细胞内低分子量的磷酰基供体——中心代谢物乙酰磷酸(Acetyl Phosphate,AcP)磷酸化[15]。RcsB形成同源二聚体或与辅助因子RcsA、BglJ、GadE、MatA等形成异源二聚体,与DNA上的Rcs结合序列结合,从而激活或抑制靶标基因的转录[4]。
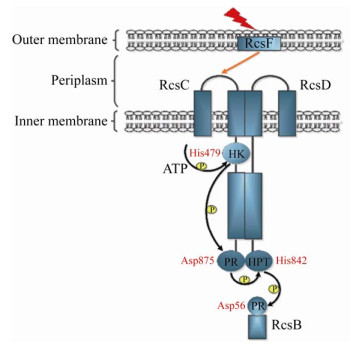
|
| 图 1 E.coli中Rcs系统的磷酸传递步骤 Figure 1 Phosphorylation of Rcs system in E.coli 注:HK:组氨酸蛋白激酶;PR:磷酸接受功能域;HPT:磷酸信号传递功能域 Note: HK: Histidine kinase; PR: Phosphoryl receiver; HPT: Histidine-containing phosphotransmitter |
|
|
目前,细菌中Rcs系统的研究重点主要在以下几个方面:Rcs系统对上游信号的感应[16-17];Rcs系统磷酸信号传递及关键氨基酸[18-19];RcsB Box及新型辅助蛋白的发现[20-21];Rcs系统所调控的下游靶基因以及生命现象的扩展[6, 11];Rcs系统与其他调控因子的作用[22-23];Rcs蛋白结构的解析等[24-25]。本文主要对Rcs系统上游信号的感知与传导、Rcs系统所调控的下游靶基因及其生命现象进行讨论。
2 Rcs系统上游信号的感知与传导细胞胞膜(Cell Envelope)是革兰氏阴性细菌特有的结构屏障,由细胞内膜(Inner Membrane)、细胞外膜(Outer Membrane)及膜之间含有肽聚糖的粘性周质空间(Periplasm)组成[9]。该结构不仅对细胞的结构完整性至关重要,而且还充当细菌与其周围环境相互作用的界面[26]。细胞胞膜不是一个静态结构,而是会根据环境的变化进行主动重建[27]。除了外部条件引发的环境信号,细菌自身膜结构变化所带来的胞膜压力也是重要的应激信号[4]。细胞胞膜压力(Cell Envelope Stress)主要由自身膜组分的生物合成、组装及转运错误以及环境胁迫导致的膜损伤引起[28]。细菌胞膜应激(Envelope Stress Responses,ESRs)对于细菌,尤其是病原菌的体外生存及毒力发挥是尤为重要的,细菌能感知胞膜损伤或缺陷并调节转录组以减轻破坏性压力[9]。Rcs系统属于胞膜应激的主要成员,Rcs系统的激活或抑制通过调节靶基因的表达水平,以减轻输入信号对细胞造成的破坏[4, 9]。例如,溶菌酶诱导Rcs系统上调了E.coli中溶菌酶抑制剂的转录[29]。鼠伤寒沙门菌(Salmonella enterica serovar Typhimurium)中的氧化应激会激活Rcs系统,从而调节dps基因的转录,进而保护细菌DNA免受感染过程中宿主活性氧(Reactive Oxygen Species,ROS)产生的损害[30]。目前报道的Rcs系统的输入信号主要是引起细胞胞膜压力的细胞外或细胞内信号[4, 9]。Rcs通路是唯一具有外膜组件(RcsF)的信号转导系统,而外膜组件对于感应几乎所有的诱导线索都是必需的[16]。因此,本小节主要综述引起细胞胞膜压力的信号分子以及RcsF如何感知胞膜压力并将信号传送到Rcs系统的下游组件。
2.1 引起胞膜压力并激活Rcs系统的信号渗透压升高是第一个被报道的能激活E.coli Rcs系统的环境信号[31],此后还发现了一些其他的有关细胞胁迫输入信号[4, 9]。如表 1所示,在E.coli中,这些细胞胁迫信号包括由溶菌酶引起的周质肽聚糖的损伤[29]、由β-内酰胺类抗生素引起的瞬时细胞壁缺陷[32]、由lolA突变引起的脂蛋白分类缺陷[33]、由周质蛋白TolA丢失引起的外膜摄动[34]、由mdoH缺失导致的膜衍生寡糖合成的缺陷[35]、由waaF缺失引起的脂多糖(Lipopolysaccharide,LPS)核心多糖的缺失[7],以及在缺少某些特定的青霉素结合蛋白PBP4、PBP5、PBP7或AmpH情况下的肽聚糖修饰[36]。除了E.coli之外,其他细菌中细胞胞膜的扰动同样能引起Rcs系统的激活。例如,甘薯茎腐病菌(Dickeya dadantii)中渗透调节周质葡聚糖(Osmoregulated Periplasmic Glucans,OPGs)浓度的变化[37]、迟缓爱德华氏菌(Edwardsiella tarda)中UDP-葡萄糖脱氢酶的缺失[38]、粘质沙雷氏菌(Serratia marcescens)中肠杆菌共同抗原(Enterobacterial Common Antigen,ECA)结构的改变[39]、S.enterica serovar Typhimurium中由氧化应激引起的外膜损伤[30]以及肠道沙门菌(Salmonella enterica)中由阳离子抗菌肽诱导的细胞膜损伤[40]等均可以调节Rcs系统的活性。总之,由这些细胞外或细胞内信号引起的胞膜压力能激活Rcs系统,进而调节Rcs系统的靶基因,以使细胞更好地适应环境或遗传的变化。
| Signal | Description | Species | Function | References |
| waaF deletion | Encoding ADP-heptose-LPS heptosyltransferase 2 | E.coli | LPS core sugar deficiency | [7] |
| Lysozyme | N-acetylmuramide glycanohydrolase | E.coli | Destruction of the periplasmic peptidoglycan | [29] |
| Active oxygen | Oxidative stress | S.enterica serovar Typhimurium | Outer membrane damage | [30] |
| β-lactam antibiotics | Chemicals repressing penicillin binding protein (PBPs) | E.coli | Inhibition of peptidoglycan formation and transition to L-form state | [32] |
| lolA mutation | Encoding lipoprotein-specific chaperone | E.coli | Defect in lipoprotein sorting | [33] |
| tolA deletion | Encoding periplasm spanning protein | E.coli | Outer membrane perturbation | [34] |
| mdoH deletion | Encoding glucosyltransferase for the synthesis of periplasmic membrane-derived oligosaccharides | E.coli | Defect in membrane-derived oligosaccharides synthesis | [35] |
| pbp4, pbp5, pbp7 or ampH deletion | Encoding penicillin-binding protein | E.coli | Modification of the peptidoglycan | [36] |
| Concentration of OPGs | Osmoregulated periplasmic glucans | D.dadantii | Sensing the osmolarity of medium | [37] |
| ugd deletion | Encoding UDP-glucose dehydrogenase | Ed.tarda | Defect in LPS, a truncated core with no O-antigen attached | [38] |
| wecG mutation | Encoding ManNAcA transferases | Se.marcescens | Alteration in the enterobacterial common antigen (ECA) structure | [39] |
| Cationic antimicrobial peptides | Binding to LPS through ionic bonds with negatively charged phosphoryl groups of the lipid A of LPS | S.enterica | Outer membrane damage | [40] |
RcsF是一种小脂蛋白(14 kD),其通过外膜上脂化的N末端膜锚定的螺旋与膜相连,在周质空间上呈现一个圆环和一个中心四链β-折叠的结构[4]。RcsF充当外界环境刺激与Rcs系统之间的中介体,通过响应不同类型的胞膜压力信号来激活Rcs信号传导系统[35]。Silhavy实验室和Collet实验室近期的研究阐明了胞膜压力通过RcsF诱导Rcs系统的机制[16, 41]。这2个团队都提出了Rcs系统通过与BamA蛋白和3个β桶状蛋白(β-Barrels,即OmpA、OmpF和OmpC)相互作用来检测胞膜压力。Cho等提出RcsF和BamA之间的相互作用在RcsF感应胞膜压力信号中起主要作用(图 2A)[16]。在没有胞膜压力的情况下,RcsF被BamA集中,并被OmpA隔离为表面暴露的蛋白质[16, 41]。在这种情况下,RcsF无法与YrfF (S.enterica serovar Typhimurium中的IgaA)相互作用来激活Rcs系统,然而在这种情况下,AcP可能充当磷酰基供体,以保持低水平的RcsB-P (图 2A);在存在胞膜压力的情况下,RcsF和BamA之间的相互作用会受到损害,因此RcsF无法传递到OmpA/OmpF/ OmpC,这使得RcsF暴露于周质空间中并自由激活Rcs系统(图 2B)[16]。该模型提出RcsF通过监测Bam复合体来感知胞膜压力[16]。Konovalova等提出了另一种模型来解释RcsF如何感知由LPS缺陷所引起的胞膜压力,在他们的模型中,RcsF与β桶状蛋白形成复合物,并通过其带正电荷的表面暴露的N末端结构域直接检测LPS分子之间的横向相互作用来调节Rcs系统的活性[41-42]。这2个模型可能代表RcsF感知不同类型压力的2种不同方式。总之,这些发现阐明了RcsF在Rcs信号转导通路中作为“哨兵”感知和传递信号的机制,这对于剖析Rcs系统的调节机制非常重要。
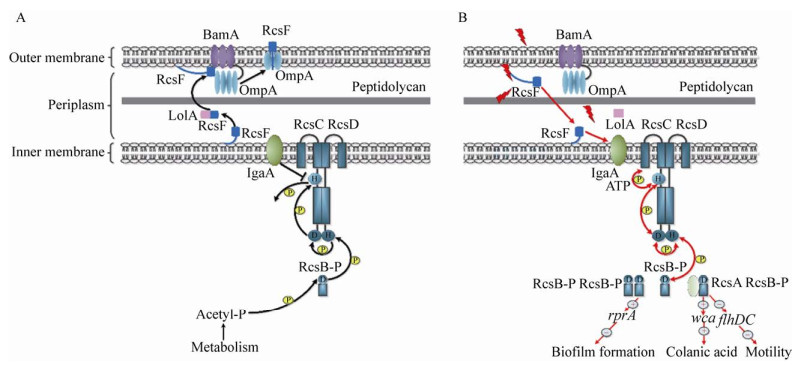
|
| 图 2 E.coli中Rcs系统的信号转导机制 Figure 2 Signal transduction of Rcs system in E.coli 注:A:在没有压力的情况下,RcsF被BamA集中,并被OmpA隔离为表面暴露的蛋白质。YrfF (S. enterica serovar Typhimurium中的IgaA)抑制了RcsC的激酶活性,从而导致RcsB去磷酸化。在这种情况下,AcP可能充当磷酰基供体,以保持低水平的RcsB-P;B:在存在压力的情况下,RcsF不与BamA和OmpA相互作用,但与IgaA相互作用,进而释放对YrfF (S. enterica serovar Typhimurium中的IgaA)的抑制作用。RcsC以ATP依赖的方式在HK域自动磷酸化。然后磷酸基团转移至RcsC的PR结构域,再转移至RcsD的HPT结构域,最后转移至RcsB。RcsB与辅助蛋白形成同源二聚体或异源二聚体,与靶基因中的保守基序结合,进而调节基因的转录 Note: A: In the absence of stress, RcsF is funneled by BamA and sequestered by OmpA as a surface-exposed protein. YrfF (IgaA in S.enterica serovar Typhimurium) inhibits the kinase activity of RcsC, resulting in dephosphorylation of RcsB. In this situation, acetyl phosphate might act as a phosphoryl group donor to keep a low level of RcsB phosphorylation. B: In the presence of stress, RcsF does not interact with BamA and OmpA but interacts with IgaA that release the inhibition of YrfF (IgaA in S.enterica serovar Typhimurium). RcsC autophosphorylates at HK domain in an ATP-dependent manner. The phosphoryl group is then transferred to the PR domain of RcsC, then to the HPT domain of RcsD and finally to RcsB. RcsB can form homodimer or heterodimers with the auxiliary proteins, which then interact with a conserved motif in target genes to modulate their transcription |
|
|
基于转录组学分析技术已成功鉴定了E.coli[43]、假结核耶尔森菌(Yersinia pseudotuberculosis)[44]、S.enterica[45]、解淀粉欧文氏菌(Erwinia amylovora)[46]、奇异变形杆菌(Proteus mirabilis)[6]、肺炎克雷伯菌(Klebsiella pneumoniae)[8]、小肠结肠炎耶尔森菌(Yersinia enterocolitica)[11]等物种中Rcs系统的基因表达谱。Rcs系统主要调节与细菌细胞表面附属结构形成相关的基因,例如荚膜、鞭毛和菌毛,以及对细胞表面维持和修饰具有预测作用的其他蛋白[12]。因此,有研究者已经提出Rcs系统将环境信号与细胞表面的重塑耦合[12]。重要的是,Rcs系统对大多数肠道细菌的毒力都具有一般的调节作用[12]。本小节将系统地综述Rcs系统调控的下游靶基因及其生命现象的扩展。
3.1 荚膜合成基因Rcs系统最初是通过其在荚膜多糖可拉酸生物合成的正调节作用中被鉴定出来。荚膜多糖可拉酸是一种细胞外多糖(Exopolysaccharide,EPS),其过量产生会导致独特的“粘液样”菌落表型[3]。实际上所有重要的荚膜多糖可拉酸其生产调节都是由Rcs系统介导[12]。Rcs系统的激活导致参与荚膜多糖可拉酸生产和运输相关基因(即cps,也称为wca操纵子)的高水平表达[12]。cps操纵子包含至少20个基因,并存在于大多数E.coli和S.enterica菌株中[47-48]。转录组学研究表明,E.coli和S.enterica这2个物种中cps操纵子的表达均受Rcs系统的调控[49-50]。Rcs系统介导的荚膜合成可调节细菌的多项生理活动[12]。研究显示E.coli中Rcs系统能感应由LPS核心多糖缺失所引发的胞膜压力,激活的Rcs系统能诱导大量荚膜多糖可拉酸的生产来促使细菌抵御外界不良环境[7]。荚膜多糖可拉酸能促进E.coli中某些菌株生物被膜的形成[51]。荚膜还与某些细菌的毒力有关。例如,在S.enterica serovar Typhimurium中,启动毒力因子Vi抗原合成所需的roiA基因被证实受Rcs系统的调控[52]。
然而某些E.coli菌株,特别是血清型O:8、O:9、O:20和O:101的菌株并不生产荚膜多糖可拉酸,而是生产其他荚膜——第1组荚膜(Group 1 Capsule),并且该荚膜也受Rcs系统的调控[53]。在对澳大利亚淡水湖中粪便大肠菌的检测中鉴定出了3种主要的E.coli菌株,研究发现这些菌株均生产第1组荚膜[54]。由于RcsA蛋白在高于30 ℃的温度下表现出不稳定性,因此荚膜多糖可拉酸通常仅在低温(< 30 ℃)下表达[12]。然而与普通E.coli菌株不同的是,鉴定出的这些菌株形成的菌落在37 ℃的琼脂平板上均呈粘液状,并且过量生产这类荚膜能促进这些菌株对淡水环境的适应[54]。综上所述,Rcs系统介导的荚膜的生产调节对肠杆菌的体外存活具有重要意义。
3.2 鞭毛和菌毛形成基因Rcs系统调控作用研究最多的是其能抑制细菌运动性和flhDC基因(编码鞭毛的主调节器)的表达。在对E.coli、S.enterica、Er.amylovora、P.mirabilis和Y.pseudotuberculosis的转录组学研究中,已经确定了Rcs系统对运动性和flhDC表达水平的负调控作用[6, 43-46]。本课题组近期的研究显示,除了flhDC,Y.enterocolitica中rcsB的突变可导致与鞭毛组装有关的12个基因(flgL、flgK、motB、motA、fliS、flgN、flgM、fliC2、fliC3、fliC、fliD和fliT)显著上调,进而导致细菌运动能力的增强[11]。据报道,在鞭毛主调节器FlhDC的帮助下,环境变化可以立即被E.coli感知,进而促使细菌朝着有利的方向移动[55]。此外,鞭毛被认为是细菌初始附着的表面细胞器,在E.coli、Y.enterocolitica以及铜绿假单胞菌(Pseudomonas aeruginosa)生物被膜的发育中起重要作用[56-58]。更重要的是,鞭毛运动是细菌活力、竞争性和致病性的重要表型特征,在E.coli和Y.enterocolitica感染的早期阶段起关键作用[59-60]。综上所述,Rcs系统对鞭毛生物合成的调节不仅限于细胞运动,还可能影响细菌的定殖,形成生物被膜甚至感染宿主的能力。
除了运动性以外,Rcs系统还抑制了其他表面附属物的产生,例如菌毛。Rcs系统可通过调节fmE和fmB基因的表达来积极调节E.coli中1型菌毛(Type 1 Fimbriae)的产生,从而使开孔细胞的频率增加[61]。在P.mirabilis中,与菌毛形成有关的mrpA、pmfA和ucaA基因均受Rcs系统的抑制[6]。菌毛虽与细菌运动无关,但菌毛能辅助细菌进行不可逆的附着,并被确定在E.coli生物被膜的发育中扮演重要角色[56]。此外,1型菌毛是限制肠炎沙门氏菌(Salmonella enterica serovar Enteritidis)在小鼠中传播和定居的重要因素,有助于S.enterica serovar Enteritidis入侵期间诱导肠道炎症反应[62]。可见,Rcs系统对菌毛生物合成的调节可能影响细菌生物被膜形成甚至侵染宿主的能力。
3.3 毒力基因Rcs系统对毒力的调节作用已在S.enterica serovar Typhimurium中进行了详尽的研究,Rcs系统对某些基因不适当地上调和/或下调会导致S.enterica毒性的减弱。据报道,rcsC等位基因(编码一种具有激酶活性的蛋白质)的突变可降低S.enterica对小鼠的毒力。该毒力的降低取决于RcsB磷酸化水平的改变,部分取决于RcsA和可拉酸的产生[63-64]。此外,激活的RcsC (由rcsC11等位基因编码)可降低鼠巨噬细胞对Salmonella的吞噬速率,并且该缺陷可能归因于可拉酸产量的增加[64]。rcsC11等位基因的激活也显著降低了Salmonella在巨噬细胞中持久的存活能力,并且该缺陷可通过突变可拉酸合成基因得以部分恢复[64]。由此可见,Salmonella中Rcs系统的激活对巨噬细胞的吞噬作用和细菌在巨噬细胞中的持久性存活都是有害的。在其他研究中,Salmonella的rcsC基因被证实在小鼠的全身性感染中起重要作用[49, 65]。BALB/c小鼠在口服rcsC突变体后,其对小鼠的毒性比口服野生型菌株要低;此外,rcsC突变体在感染小鼠11 d后,小鼠脾脏和肝脏的恢复期较口服野生型菌株显著降低[65]。
随后的微阵列研究支持了一个观点,即Rcs系统可能参与了Salmonella毒力发挥期间基因表达的时间调控。研究表明,Rcs系统可以正调节(低RcsB-P水平)或负调节(高RcsB-P水平) Salmonella中SPI-1和SPI-2致病岛基因的表达水平[66-67]。SPI-1和SPI-2致病岛在细菌感染的早期阶段很重要,可促进S.enterica穿过上皮屏障并进入巨噬细胞[68-69]。图 3A很好地解释了Salmonella感染过程中Rcs系统如何对毒力基因的表达水平进行时间上的控制:在感染的早期阶段,细胞中RcsB-P水平较低,从而允许表达与运动、SPI-1和SPI-2致病岛相关的基因。在Salmonella进入巨噬细胞后,环境信号触发了磷酸激酶活性的激活,从而导致细胞中RcsB-P的水平增加,进而抑制了运动、SPI-1和SPI-2致病岛相关基因的表达,同时驱动全身性感染所需基因的表达[12]。此时,Rcs系统正调控ydeI的表达,该基因编码14 kD的周质蛋白,对S.enterica serovar Typhimurium持续感染小鼠很重要[49, 70]。该模型表明,Rcs系统在S.enterica毒力发挥的时间调控中起作用,并且该系统的激活参与驱动从局部感染向全身感染的转变,该模型类似于Rcs系统控制E.coli生物被膜发育的模型(图 3B);在该模型中,RcsB-P水平的升高可通过抑制运动性和诱导EPS的产生,进而促使细菌从起始附着阶段向形成成熟生物被膜阶段进行转变[71]。

|
| 图 3 Rcs系统对基因表达进行时间控制的模型 Figure 3 Model for the temporal control of gene expression by the Rcs system 注:A:当S.enterica serovar Typhimurium进入巨噬细胞后,环境信号会触发Rcs系统的激活。RcsB-P水平的升高抑制了细菌粘附和感染巨噬细胞所需基因(运动、SPI-1和SPI-2致病岛相关基因)的表达,同时驱动全身感染所需基因(ydeI)的表达;B:在E.coli中,Rcs系统的激活发生在细菌附着到固体表面后的某个时刻。RcsB-P水平的升高会抑制细菌附着所需基因(运动和卷曲菌毛合成相关基因)的表达,并诱导EPS (负责成熟生物被膜形成)合成基因的表达 Note: A: After S.enterica serovar Typhimurium has gained entry into the macrophage, environmental signals trigger the activation of Rcs system. The increase in RcsB-P levels inhibits the expression of genes required for bacterial adhesion and infection (motility, SPI-1 and SPI-2) to macrophages, while driving the expression of genes required for systemic infection (ydeI); B: In E.coli, the activation of Rcs system occurs at some point after attachment to a solid surface. The increase in RcsB-P levels represses the expression of genes required for attachment (motility and curli) and induces the expression of genes responsible for the production of EPS, which are required for the development of a mature biofilm |
|
|
此外,Rcs系统已被证实可调节肠出血性大肠杆菌(Enterohemorrhagic E.coli,EHEC)中LEE (Locus of Enterocyte Effacement)致病岛基因的表达,LEE位点编码组装Ⅲ型分泌系统(Type Ⅲ Secretion System,T3SS)所需的所有蛋白;在该菌中,Rcs系统的激活可通过正向调节grvA的表达而促进LEE基因的转录,致使T3SS效应蛋白的表达量上调,最终导致细菌对宿主细胞的黏附和侵染能力增强[72]。在Y.pseudotuberculosis中,Rcs系统可通过调控virG-lcrF操纵子(编码LcrF)的转录而正向调控Ysc-Yop T3SS,使细菌在宿主免疫细胞中分泌的Yop效应蛋白增加,从而增强细菌的存活能力,临床上可造成败血症等严重感染[73]。在Y.enterocolitica中,rcsB的缺失可显著下调Ysa T3SS的表达,Ysa T3SS参与Y.enterocolitica在小鼠末端回肠的定殖,当将野生型Y.enterocolitica与rcsB突变体共同感染小鼠时,rcsB突变体在适应性上表现出了劣势[74-75]。本课题组近期的研究表明,rcsB的缺失能激活Y.enterocolitica对Caco-2细胞的黏附和侵染能力,这可能是由于rcsB突变体激活了鞭毛合成和细菌趋化行为所导致的结果[11]。可见,Rcs系统对Y.enterocolitica毒力的调控存在体内和体外的差异。除了这些物种之外,Rcs系统也被证实能在植物病原菌Er.amylovora和果胶杆菌(Pectobacterium carotovorum)的毒力调控中起作用[76-77]。因此,尽管Rcs系统对毒力的调控在S.enterica中进行了详尽的研究,但该系统对整个肠道细菌的毒力可能具有一般的调节作用。
3.4 σ因子在E.coli中,研究发现Rcs系统能在转录后水平上调节基因的表达。研究表明E.coli中的RprA (一种sRNA)受Rcs系统的正调控,并且该sRNA能与rpoS mRNA (编码选择性σ因子,σs)的5′末端进行碱基配对,从而提高该mRNA的翻译水平[78-79]。Rcs系统的激活可导致E.coli中RprA的表达量增加50-60倍,进而导致细胞中σs蛋白的表达水平增加32-35倍[78]。Rcs系统还可调节E.coli中lrhA的表达水平,该基因编码LysR型转录调控因子[80]。LrhA通过调控一个未知的sRNA来促进rpoS mRNA转录本的转录后调控[80]。可见,Rcs系统的主要监管目标之一似乎是σs。Rcs系统可通过调控σs的表达水平来间接调控细菌的生理行为。例如,与rcsC或rcsD突变有关的E.coli生物被膜形成缺陷是由于在这些基因型背景下细胞中σs的表达水平升高所导致,进一步发现该生物被膜形成缺陷可通过突变rcsB或rprA得以恢复[81]。据报道,σs表达水平的提高可通过σ因子竞争过程来降低某些受σ70调控基因的表达,并且有证据表明σs可通过该过程抑制cps操纵子和yjbEFGH操纵子的表达,这2个操纵子都编码同一个EPS且都受Rcs系统的正调控[82-83]。
Rcs系统的特征是许多(但不是全部)受该系统调控基因的表达是瞬时的。当Rcs途径被激活后基因的表达水平迅速增加,随后表达量下降至新的稳态水平[31, 50]。实现该瞬时表达谱的机制是未知的,或许由Rcs系统和σs形成的监管网络可以解释这种现象,预计Rcs系统和σs会形成一种称为非相干1型前馈环(Incoherent Type 1 Feedforward LoopI,I1-FFL)的网络基序,I1-FFL基序是脉冲发生器,当Rcs系统被激活后(例如渗透压胁迫),预计受该基序控制的基因(例如cps操纵子)会瞬时表达,进而允许细胞通过产生诸如可拉酸等物质来促使细胞本能地响应对细菌具有潜在致命性的环境压力;然后这种“本能”或“反馈”响应会在cps的表达过程中出现依赖于σs的调控机制,使其表达水平更加适应外界环境[84]。可见,E.coli中的Rcs系统似乎是一个复杂的调控网络,涉及转录和转录后水平的基因调控。
3.5 其他基因随着对Rcs系统的不断研究,越来越多的基因被鉴定出受该系统的调控。研究表明与细胞分裂有关的ftsAZ操纵子在E.coli和P.mirabilis中被RcsB激活,然而在Y.enterocolitica中则被RcsB抑制[6, 11, 85]。本课题组近期的研究发现,Y.enterocolitica中rcsB的突变上调了大量与细菌趋化信号转导系统相关的基因(例如mcp、cheA和cheY等)[11]。在P.mirabilis中,与趋化性相关的基因cheB、cheR和cheY也被鉴定为受RcsB抑制[6]。Rcs系统还抑制了鼠疫耶尔森菌(Yersinia pestis)和Y.enterocolitica中与胞外多糖聚β-1, 6-N-乙酰氨基葡萄糖和胞内第二信使分子c-di-GMP生物合成所需基因hmsHFRS和hmsT的表达,进而抑制细菌生物被膜的形成[86-87]。Rcs系统还可调节细菌对阳离子抗菌肽的抗性。已显示S.enterica serovar Typhimurium中RcsC能调节ugd的表达水平,该基因负责L-氨基阿拉伯糖合成和掺入LPS,以诱导细菌对多粘菌素B的抗性[88]。本课题组前期研究已经确定了Y.enterocolitica中RcsB对抗多粘菌素B的积极作用[87]。随后的转录组学分析显示rcsB的缺失导致与多粘菌素B抗性相关phoQ和pagP基因的表达显著下调,pagP是PhoP/PhoQ双组分调节系统的靶标基因,负责LPS结构的修饰以提高细菌对多粘菌素B的抗性,这些发现使我们推测Rcs系统通过正向调节PhoP/PhoQ系统的活性,从而赋予Y.enterocolitica对多粘菌素B一定水平的抗性[11]。细菌中受Rcs系统调节的基因见表 2。
| Gene | Function | Species | References |
| flhDC | Encoding the master regulator of flagella | E.coli, S.enterica, Er.amylovora, P.mirabilis, Y.pseudotuberculosis, Y.enterocolitica | [6, 11, 43-46] |
| cheY | Encoding chemotactic response regulator | P.mirabilis, Y.enterocolitica | [6, 11] |
| ydeI | Increasing resistance to antimicrobial peptides | S.enterica serovar Typhimurium | [49] |
| cps | Responsible for the synthesis of colanic acid | E.coli, S.enterica | [49-50] |
| roiA | Initiating Vi antigen production | S.enterica serovar Typhimurium | [52] |
| fmE, fmB; mrpA, pmfA, ucaA | Responsible for the synthesis of fimbriae | E.coli; P.mirabilis | [6, 61] |
| SPI-1 and SPI-2 pathogenicity islands | Encoding virulence factor | S.enterica serovar Typhimurium | [66-67] |
| LEE pathogenicity island | Encoding proteins required for assembly of T3SS | EHEC | [72] |
| lcrF | Regulating the expression of Ysc-Yop T3SS | Y.pseudotuberculosis | [73] |
| rprA | Responsible for regulating the expression level of σs | E.coli | [78] |
| lrhA | Responsible for regulating the expression level of σs | E.coli | [80] |
| ftsAZ | Encoding cell division-associated protein | E.coli, P.mirabilis, Y.enterocolitica | [6, 11, 85] |
| hmsHFRS, hmsT | Responsible for the synthesis of poly-β-1, 6-N-acetylglucosamine EPS and intracellular c-di-GMP | Y.pestis, Y. enterocolitica | [86-87] |
| ugd; pagP | Responsible for LPS modification to increase resistance to polymyxin B | S.enterica serovar Typhimurium; Y.enterocolitica | [11, 88] |
Rcs双组分调节系统是仅在肠杆菌科细菌中发现的重要信号转导途径,该系统能整合环境信号,调节基因表达并改变细菌的生理行为。外膜蛋白RcsF能感知激活Rcs系统的信号分子(引起胞膜压力的细胞外或细胞内信号),触发Rcs系统下游信号的传递:RcsC→RcsD→RcsB,最后对靶基因进行调控。RcsC的双重功能、RcsB的磷酸化修饰以及多种辅助蛋白的参与共同导致Rcs系统的复杂性和灵活性,从而实现对其目标基因的精确调控。肠杆菌科特有的Rcs系统可调控荚膜合成、鞭毛和菌毛形成、σ因子、抗性基因表达等而抵抗外界环境压力,也可调控细菌毒力。
本文总结了Rcs系统上游信号的感知与传导、Rcs系统调节的下游靶基因及其生命现象,可以看出Rcs系统在细菌环境应答方面扮演重要角色。尽管目前对于细菌Rcs系统环境应答机制的研究取得了重要的进展,仍然还存在着许多亟待解决的科学问题:(1)在没有RcsF的情况下,Rcs系统似乎可以正常工作,而Rcs系统如何感知信号分子仍然未知。(2) Rcs系统环境响应机制的研究主要集中在2个方向,即激活Rcs系统的特定环境信号分子和受Rcs系统调控的下游靶基因。但是,Rcs系统与其他TCS之间的作用机制尚不清楚。例如,Rcs系统如何与PhoP/PhoQ系统相互作用以调节Y.enterocolitica对多粘菌素B的抗性?(3)截至目前,大多数研究都是在体外生长条件下或通过过度激活Rcs系统进行研究,很少有研究在体内条件下对Rcs系统的基因表达谱进行解析。由于Rcs系统对基因的表达调控存在体内和体外的差异,因此,有必要进行体内Rcs系统的转录组学研究,以寻找新的毒力相关靶标。总而言之,Rcs系统为研究细菌环境适应的复杂性提供了一个独特的模型,对Rcs系统的理解有助于为细菌的控制、预防及治疗方案的开发提供理论依据。
| [1] |
Wall E, Majdalani N, Gottesman S. The complex Rcs regulatory cascade[J]. Annual Review of Microbiology, 2018, 72(1): 111-139. DOI:10.1146/annurev-micro-090817-062640 |
| [2] |
Hoch JA. Two-component and phosphorelay signal transduction[J]. Current Opinion in Microbiology, 2000, 3(2): 165-170. DOI:10.1016/S1369-5274(00)00070-9 |
| [3] |
Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes[J]. Journal of Bacteriology, 1985, 162(3): 1111-1119. DOI:10.1128/JB.162.3.1111-1119.1985 |
| [4] |
Guo XP, Sun YC. New insights into the non-orthodox two component Rcs phosphorelay system[J]. Frontiers in Microbiology, 2017, 8: 2014. DOI:10.3389/fmicb.2017.02014 |
| [5] |
Clarke EJ, Voigt CA. Characterization of combinatorial patterns generated by multiple two-component sensors in E.coli that respond to many stimuli[J]. Biotechnology and Bioengineering, 2011, 108(3): 666-675. DOI:10.1002/bit.22966 |
| [6] |
Howery KE, Clemmer KM, Rather PN. The Rcs regulon in Proteus mirabilis: implications for motility, biofilm formation, and virulence[J]. Current Genetics, 2016, 62(4): 775-789. DOI:10.1007/s00294-016-0579-1 |
| [7] |
Ren G, Wang Z, Li Y, Hu XQ, Wang XY. Effects of lipopolysaccharide core sugar deficiency on colanic acid biosynthesis in Escherichia coli[J]. Journal of Bacteriology, 2016, 198(11): 1576-1584. DOI:10.1128/JB.00094-16 |
| [8] |
Su KW, Peng D, Liu P, Zhang ZS, Li X, He Q, Qiu JF, Li YL. Identification of RcsB transcriptional profile in Klebsiella pneumoniae by microarray[J]. Genomics and Applied Biology, 2018, 37(8): 3611-3618. (in Chinese) 苏克文, 彭丹, 刘品, 张仲双, 李璇, 何蔷, 邱景富, 李迎丽. 基因芯片技术探究肺炎克雷伯菌RcsB的转录谱[J]. 基因组学与应用生物学, 2018, 37(8): 3611-3618. |
| [9] |
Mitchell AM, Silhavy TJ. Envelope stress responses: balancing damage repair and toxicity[J]. Nature Reviews Microbiology, 2019, 17(7): 417-428. DOI:10.1038/s41579-019-0199-0 |
| [10] |
Meng J, Huang C, Huang XN, Liu DY, Han BZ, Chen JY. Osmoregulated periplasmic glucans transmit external signals through Rcs phosphorelay pathway in Yersinia enterocolitica[J]. Frontiers in Microbiology, 2020, 11: 122. DOI:10.3389/fmicb.2020.00122 |
| [11] |
Meng J, Bai JQ, Chen JY. Transcriptomic analysis reveals the role of RcsB in suppressing bacterial chemotaxis, flagellar assembly and infection in Yersinia enterocolitica[J]. Current Genetics, 2020, 66(5): 971-988. DOI:10.1007/s00294-020-01083-x |
| [12] |
Clarke DJ. The Rcs phosphorelay: more than just a two-component pathway[J]. Future Microbiology, 2010, 5(8): 1173-1184. DOI:10.2217/fmb.10.83 |
| [13] |
Pristovšek P, Sengupta K, Löhr F, Schäfer B, von Trebra MW, Rüterjans H, Bernhard F. Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box[J]. Journal of Biological Chemistry, 2003, 278(20): 17752-17759. DOI:10.1074/jbc.M301328200 |
| [14] |
Hussein NA, Cho SH, Laloux G, Siam R, Collet JF. Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the Rcs phosphorelay across subcellular compartments[J]. PLoS Genetics, 2018, 14(5). |
| [15] |
Hu LD, Chi BK, Kuhn ML, Filippova EV, Walker-Peddakotla AJ, Bäsell K, Becher D, Anderson WF, Antelmann H, Wolfe AJ. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter[J]. Journal of Bacteriology, 2013, 195(18): 4174-4186. DOI:10.1128/JB.00383-13 |
| [16] |
Cho SH, Szewczyk J, Pesavento C, Zietek M, Banzhaf M, Roszczenko P, Asmar A, Laloux G, Hov AK, Leverrier P, et al. Detecting envelope stress by monitoring β-barrel assembly[J]. Cell, 2014, 159(7): 1652-1664. DOI:10.1016/j.cell.2014.11.045 |
| [17] |
Sato T, Takano A, Hori N, Izawa T, Eda T, Sato K, Umekawa M, Miyagawa H, Matsumoto K, Muramatsu-Fujishiro A, et al. Role of the inner-membrane histidine kinase RcsC and outer-membrane lipoprotein RcsF in the activation of the Rcs phosphorelay signal transduction system in Escherichia coli[J]. Microbiology, 2017, 163(7): 1071-1080. DOI:10.1099/mic.0.000483 |
| [18] |
Clarke DJ, Joyce SA, Toutain CA, Jacq A, Holland IB. Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12[J]. Journal of Bacteriology, 2002, 184(4): 1204-1208. DOI:10.1128/jb.184.4.1204-1208.2002 |
| [19] |
Ancona V, Chatnaparat T, Zhao YF. Conserved aspartate and lysine residues of RcsB are required for amylovoran biosynthesis, virulence, and DNA binding in Erwinia amylovora[J]. Molecular Genetics and Genomics, 2015, 290(4): 1265-1276. DOI:10.1007/s00438-015-0988-8 |
| [20] |
Su KW, Zhou XP, Luo M, Xu X, Liu P, Li X, Xue J, Chen S, Xu W, Li YL, et al. Genome-wide identification of genes regulated by RcsA, RcsB, and RcsAB phosphorelay regulators in Klebsiella pneumoniae NTUH-K2044[J]. Microbial Pathogenesis, 2018, 123: 36-41. DOI:10.1016/j.micpath.2018.06.036 |
| [21] |
Salscheider SL, Jahn A, Schnetz K. Transcriptional regulation by BglJ-RcsB, a pleiotropic heteromeric activator in Escherichia coli[J]. Nucleic Acids Research, 2014, 42(5): 2999-3008. DOI:10.1093/nar/gkt1298 |
| [22] |
Winfield MD, Latifi T, Groisman EA. Transcriptional regulation of the 4-amino-4-deoxy-L-arabinose biosynthetic genes in Yersinia pestis[J]. Journal of Biological Chemistry, 2005, 280(15): 14765-14772. DOI:10.1074/jbc.M413900200 |
| [23] |
Stratmann T, Pul U, Wurm R, Wagner R, Schnetz K. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants[J]. Molecular Microbiology, 2012, 83(6): 1109-1123. DOI:10.1111/j.1365-2958.2012.07993.x |
| [24] |
Schmöe K, Rogov VV, Rogova NY, Löhr F, Güntert P, Bernhard F, Dötschi V. Structural insights into Rcs phosphotransfer: the newly identified RcsD-ABL domain enhances interaction with the response regulator RcsB[J]. Structure, 2011, 19(4): 577-587. DOI:10.1016/j.str.2011.01.012 |
| [25] |
Filippova EV, Wawrzak Z, Ruan JP, Pshenychnyi S, Schultz RM, Wolfe AJ, Anderson WF. Crystal structure of nonphosphorylated receiver domain of the stress response regulator RcsB from Escherichia coli[J]. Protein Science, 2016, 25(12): 2216-2224. DOI:10.1002/pro.3050 |
| [26] |
Rojas ER, Billings G, Odermatt PD, Auer GK, Zhu LL, Miguel A, Chang F, Weibel DB, Theriot JA, Huang KC. The outer membrane is an essential load-bearing element in Gram-negative bacteria[J]. Nature, 2018, 559(7715): 617-621. DOI:10.1038/s41586-018-0344-3 |
| [27] |
Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope[J]. Cold Spring Harbor Perspectives in Biology, 2010, 2(5): a000414. |
| [28] |
Grabowicz M, Silhavy TJ. Envelope stress responses: an interconnected safety net[J]. Trends in Biochemical Sciences, 2017, 42(3): 232-242. DOI:10.1016/j.tibs.2016.10.002 |
| [29] |
Callewaert L, Vanoirbeek KGA, Lurquin I, Michiels CW, Aertsen A. The Rcs two-component system regulates expression of lysozyme inhibitors and is induced by exposure to lysozyme[J]. Journal of Bacteriology, 2009, 191(6): 1979-1981. DOI:10.1128/JB.01549-08 |
| [30] |
Farizano JV, Torres MA, de las Mercedes Pescaretti M, Delgado MA. The RcsCDB regulatory system plays a crucial role in the protection of Salmonella enterica serovar Typhimurium against oxidative stress[J]. Microbiology, 2014, 160(10): 2190-2199. DOI:10.1099/mic.0.081133-0 |
| [31] |
Sledjeski DD, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12[J]. Journal of Bacteriology, 1996, 178(4): 1204-1206. DOI:10.1128/JB.178.4.1204-1206.1996 |
| [32] |
Hirakawa H, Nishino K, Yamada J, Hirata T, Yamaguchi A. β-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli[J]. Journal of Antimicrobial Chemotherapy, 2003, 52(4): 576-582. DOI:10.1093/jac/dkg406 |
| [33] |
Tao K, Narita SI, Tokuda H. Defective lipoprotein sorting induces lolA expression through the Rcs stress response phosphorelay system[J]. Journal of Bacteriology, 2012, 194(14): 3643-3650. DOI:10.1128/JB.00553-12 |
| [34] |
Morgan JK, Ortiz JA, Riordan JT. The role for TolA in enterohemorrhagic Escherichia coli pathogenesis and virulence gene transcription[J]. Microbial Pathogenesis, 2014, 77: 42-52. DOI:10.1016/j.micpath.2014.10.010 |
| [35] |
Shiba Y, Miyagawa H, Nagahama H, Matsumoto K, Kondo D, Matsuoka S, Matsumoto K, Hara H. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli[J]. Microbiology, 2012, 158(5): 1238-1248. DOI:10.1099/mic.0.056945-0 |
| [36] |
Evans KL, Kannan S, Li G, de Pedro MA, Young KD. Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli[J]. Journal of Bacteriology, 2013, 195(19): 4415-4424. DOI:10.1128/JB.00596-13 |
| [37] |
Bontemps-Gallo S, Madec E, Dondeyne J, Delrue B, Robbe-Masselot C, Vidal O, Prouvost AF, Boussemart G, Bohin JP, Lacroix JM. Concentration of osmoregulated periplasmic glucans (OPGs) modulates the activation level of the RcsCD RcsB phosphorelay in the phytopathogen bacteria Dickeya dadantii[J]. Environmental Microbiology, 2013, 15(3): 881-894. DOI:10.1111/1462-2920.12054 |
| [38] |
Lv YZ, Zheng JY, Yang MJ, Wang QY, Zhang YX. An Edwardsiella tarda mutant lacking UDP-glucose dehydrogenase shows pleiotropic phenotypes, attenuated virulence, and potential as a vaccine candidate[J]. Veterinary Microbiology, 2012, 160(3/4): 506-512. |
| [39] |
Castelli ME, Véscovi EG. The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens[J]. Journal of Bacteriology, 2011, 193(1): 63-74. DOI:10.1128/JB.00839-10 |
| [40] |
Farris C, Sanowar S, Bader MW, Pfuetzner R, Miller SI. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF[J]. Journal of Bacteriology, 2010, 192(19): 4894-4903. DOI:10.1128/JB.00505-10 |
| [41] |
Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins[J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(41): E4350-E4358. DOI:10.1073/pnas.1417138111 |
| [42] |
Konovalova A, Mitchell AM, Silhavy TJ. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane[J]. eLife, 2016, 5: e15276. DOI:10.7554/eLife.15276 |
| [43] |
Hagiwara D, Sugiura M, Oshima T, Mori H, Aiba H, Yamashino T, Mizuno T. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli[J]. Journal of Bacteriology, 2003, 185(19): 5735-5746. DOI:10.1128/JB.185.19.5735-5746.2003 |
| [44] |
Hinchliffe SJ, Howard SL, Huang YH, Clarke DJ, Wren BW. The importance of the Rcs phosphorelay in the survival and pathogenesis of the enteropathogenic Yersiniae[J]. Microbiology, 2008, 154(4): 1117-1131. DOI:10.1099/mic.0.2007/012534-0 |
| [45] |
Mariscotti JF, García-del Portillo F. Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA[J]. Journal of Bacteriology, 2009, 191(6): 1855-1867. DOI:10.1128/JB.01604-08 |
| [46] |
Wang DP, Qi MS, Calla B, Korban SS, Clough SJ, Cock PJA, Sundin GW, Toth I, Zhao YF. Genome-wide identification of genes regulated by the Rcs phosphorelay system in Erwinia amylovora[J]. Molecular Plant-Microbe Interactions, 2012, 25(1): 6-17. DOI:10.1094/MPMI-08-11-0207 |
| [47] |
Rahn A, Drummelsmith J, Whitfield C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae[J]. Journal of Bacteriology, 1999, 181(7): 2307-2313. DOI:10.1128/JB.181.7.2307-2313.1999 |
| [48] |
Stevenson G, Lan R, Reeves PR. The colanic acid gene cluster of Salmonella enterica has a complex history[J]. FEMS Microbiology Letters, 2000, 191(1): 11-16. DOI:10.1111/j.1574-6968.2000.tb09312.x |
| [49] |
Erickson KD, Detweiler CS. The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice[J]. Molecular Microbiology, 2006, 62(3): 883-894. DOI:10.1111/j.1365-2958.2006.05420.x |
| [50] |
Ferrières L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface[J]. Molecular Microbiology, 2003, 50(5): 1665-1682. DOI:10.1046/j.1365-2958.2003.03815.x |
| [51] |
Danese PN, Pratt LA, Kolter R. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture[J]. Journal of Bacteriology, 2000, 182(12): 3593-3596. DOI:10.1128/JB.182.12.3593-3596.2000 |
| [52] |
Arricau N, Hermant D, Waxin H, Ecobichon C, Duffey PS, Popoff MY. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity[J]. Molecular Microbiology, 1998, 29(3): 835-850. DOI:10.1046/j.1365-2958.1998.00976.x |
| [53] |
Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli[J]. Molecular Microbiology, 1999, 31(5): 1307-1319. DOI:10.1046/j.1365-2958.1999.01276.x |
| [54] |
Power ML, Littlefield-Wyer J, Gordon DM, Veal DA, Slade MB. Phenotypic and genotypic characterization of encapsulated Escherichia coli isolated from blooms in two Australian lakes[J]. Environmental Microbiology, 2005, 7(5): 631-640. DOI:10.1111/j.1462-2920.2005.00729.x |
| [55] |
Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli[J]. Journal of General and Applied Microbiology, 1967, 46(2): 175-184. DOI:10.1099/00221287-46-2-175 |
| [56] |
van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation[J]. Research in Microbiology, 2005, 156(5/6): 626-633. |
| [57] |
Meng J, Xu JG, Chen JY. The role of osmoregulated periplasmic glucans in the biofilm antibiotic resistance of Yersinia enterocolitica[J]. Microbial Pathogenesis, 2020, 147: 104284. DOI:10.1016/j.micpath.2020.104284 |
| [58] |
Liaqat I, Liaqat M, Tahir HM, Ikram-ul-Haq, Ali NM, Arshad M, Arshad N. Motility effects biofilm formation in Pseudomonas aeruginosa and Enterobacter cloacae[J]. Pakistan Journal of Pharmaceutical Sciences, 2019, 32(3): 927-932. |
| [59] |
Josenhans C, Suerbaum S. The role of motility as a virulence factor in bacteria[J]. International Journal of Medical Microbiology, 2002, 291(8): 605-614. DOI:10.1078/1438-4221-00173 |
| [60] |
Young GM, Badger JL, Miller VL. Motility is required to initiate host cell invasion by Yersinia enterocolitica[J]. Infection and Immunity, 2000, 68(7): 4323-4326. DOI:10.1128/IAI.68.7.4323-4326.2000 |
| [61] |
Schwan WR, Shibata S, Aizawa SI, Wolfe AJ. The two-component response regulator RcsB regulates type 1 piliation in Escherichia coli[J]. Journal of Bacteriology, 2007, 189(19): 7159-7163. DOI:10.1128/JB.00705-07 |
| [62] |
Kuźmińska-Bajor M, Grzymajło K, Ugorski M. Type 1 fimbriae are important factors limiting the dissemination and colonization of mice by Salmonella enteritidis and contribute to the induction of intestinal inflammation during Salmonella invasion[J]. Frontiers in Microbiology, 2015, 6: 276. |
| [63] |
García-Calderón CB, García-Quintanilla M, Casadesús J, Ramos-Morales F. Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system[J]. Microbiology, 2005, 151(2): 579-588. DOI:10.1099/mic.0.27520-0 |
| [64] |
Mouslim C, Delgado M, Groisman EA. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence[J]. Molecular Microbiology, 2004, 54(2): 386-395. DOI:10.1111/j.1365-2958.2004.04293.x |
| [65] |
Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance[J]. Molecular Microbiology, 2003, 48(2): 385-400. DOI:10.1046/j.1365-2958.2003.03455.x |
| [66] |
Wang QF, Zhao YF, McClelland M, Harshey RA. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: Dual regulation of flagellar and SPI-2 virulence genes[J]. Journal of Bacteriology, 2007, 189(23): 8447-8457. DOI:10.1128/JB.01198-07 |
| [67] |
Wang QF, Harshey RM. Rcs signaling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility[J]. Molecular Microbiology, 2009, 74(1): 71-84. DOI:10.1111/j.1365-2958.2009.06851.x |
| [68] |
Lostroh CP, Lee CA. The Salmonella pathogenicity island-1 type Ⅲ secretion system[J]. Microbes and Infection, 2001, 3(14/15): 1281-1291. |
| [69] |
Fass E, Groisman EA. Control of Salmonella pathogenicity island-2 gene expression[J]. Current Opinion in Microbiology, 2009, 12(2): 199-204. DOI:10.1016/j.mib.2009.01.004 |
| [70] |
Pilonieta MC, Erickson KD, Ernst RK, Detweiler CS. A protein important for antimicrobial peptide resistance, YdeI/OmdA, is in the periplasm and interacts with OmpD/NmpC[J]. Journal of Bacteriology, 2009, 191(23): 7243-7252. DOI:10.1128/JB.00688-09 |
| [71] |
Huang YH, Ferrières L, Clarke DJ. The role of the Rcs phosphorelay in Enterobacteriaceae[J]. Research in Microbiology, 2006, 157(3): 206-212. DOI:10.1016/j.resmic.2005.11.005 |
| [72] |
Tobe T, Ando H, Ishikawa H, Abe H, Tashiro K, Hayashi T, Kuhara S, Sugimoto N. Dual regulatory pathways integrating the RcsC-RcsD-RcsB signalling system control enterohaemorrhagic Escherichia coli pathogenicity[J]. Molecular Microbiology, 2005, 58(1): 320-333. DOI:10.1111/j.1365-2958.2005.04828.x |
| [73] |
Li YL, Hu YB, Francis MS, Chen SY. RcsB positively regulates the Yersinia Ysc-Yop type Ⅲ secretion system by activating expression of the master transcriptional regulator LcrF[J]. Environmental Microbiology, 2015, 17(4): 1219-1233. DOI:10.1111/1462-2920.12556 |
| [74] |
Venecia K, Young GM. Environmental regulation and virulence attributes of the Ysa type Ⅲ secretion system of Yersinia enterocolitica biovar 1B[J]. Infection and Immunity, 2005, 73(9): 5961-5977. DOI:10.1128/IAI.73.9.5961-5977.2005 |
| [75] |
Walker KA, Miller VL. Synchronous gene expression of the Yersinia enterocolitica Ysa type Ⅲ secretion system and its effectors[J]. Journal of Bacteriology, 2009, 191(6): 1816-1826. DOI:10.1128/JB.01402-08 |
| [76] |
Wang DP, Korban SS, Zhao YF. The Rcs phosphorelay system is essential for pathogenicity in Erwinia amylovora[J]. Molecular Plant Pathology, 2009, 10(2): 277-290. DOI:10.1111/j.1364-3703.2008.00531.x |
| [77] |
Andresen L, Kõiv V, Alamäe T, Mäe A. The Rcs phosphorelay modulates the expression of plant cell wall degrading enzymes and virulence in Pectobacterium carotovorum ssp. carotovorum[J]. FEMS Microbiology Letters, 2007, 273(2): 229-238. DOI:10.1111/j.1574-6968.2007.00794.x |
| [78] |
Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA[J]. Molecular Microbiology, 2002, 46(3): 813-826. DOI:10.1046/j.1365-2958.2002.03203.x |
| [79] |
Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA[J]. Molecular Microbiology, 2001, 39(5): 1382-1394. DOI:10.1111/j.1365-2958.2001.02329.x |
| [80] |
Peterson CN, Carabetta VJ, Chowdhury T, Silhavy TJ. LrhA regulates rpoS translation in response to the Rcs phosphorelay system in Escherichia coli[J]. Journal of Bacteriology, 2006, 188(9): 3175-3181. DOI:10.1128/JB.188.9.3175-3181.2006 |
| [81] |
Ferrières L, Thompson A, Clarke DJ. Elevated levels of σs inhibit biofilm formation in Escherichia coli: a role for the Rcs phosphorelay[J]. Microbiology, 2009, 155(11): 3544-3553. DOI:10.1099/mic.0.032722-0 |
| [82] |
Ionescu M, Belkin S. Overproduction of exopolysaccharides by an Escherichia coli K-12 rpoS mutant in response to osmotic stress[J]. Applied and Environmental Microbiology, 2009, 75(2): 483-492. DOI:10.1128/AEM.01616-08 |
| [83] |
Ferrières L, Aslam SN, Cooper RM, Clarke DJ. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production[J]. Microbiology, 2007, 153(4): 1070-1080. DOI:10.1099/mic.0.2006/002907-0 |
| [84] |
Alon U. Network motifs: theory and experimental approaches[J]. Nature Reviews Genetics, 2007, 8(6): 450-461. DOI:10.1038/nrg2102 |
| [85] |
Hill NS, Buske PJ, Shi Y, Levin PA. A moonlighting enzyme links Escherichia coli cell size with central metabolism[J]. PLoS Genetics, 2013, 9(7): e1003663. DOI:10.1371/journal.pgen.1003663 |
| [86] |
Fang N, Yang HY, Fang HH, Liu L, Zhang YQ, Wang L, Han YP, Zhou DS, Yang RF. RcsAB is a major repressor of Yersinia biofilm development through directly acting on hmsCDE, hmsT, and hmsHFRS[J]. Scientific Reports, 2015, 5(1): 9566. DOI:10.1038/srep09566 |
| [87] |
Meng J, Bai JQ, Xu JH, Huang C, Chen JY. Differential regulation of physiological activities by RcsB and OmpR in Yersinia enterocolitica[J]. FEMS Microbiology Letters, 2019, 336(17): fnz210. |
| [88] |
Mouslim C, Groisman EA. Control of the Salmonella ugd gene by three two-component regulatory systems[J]. Molecular Microbiology, 2003, 47(2): 335-344. DOI:10.1046/j.1365-2958.2003.03318.x |
 2021, Vol. 48
2021, Vol. 48




