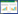扩展功能
文章信息
- 刘新育, 谢夏, 朱东东, 陈红歌
- LIU Xin-Yu, XIE Xia, ZHU Dong-Dong, CHEN Hong-Ge
- 木聚糖酶与XIP型木聚糖酶抑制蛋白相互作用分子机制的研究进展
- Interaction between xylanase and XIP-type xylanase inhibitor protein: a review
- 微生物学通报, 2020, 47(7): 2300-2308
- Microbiology China, 2020, 47(7): 2300-2308
- DOI: 10.13344/j.microbiol.china.200059
-
文章历史
- 收稿日期: 2020-01-19
- 网络首发日期: 2020-05-21
β-1, 4-内切木聚糖酶(EC 3.2.1.8)在木聚糖的降解过程中具有重要作用,其可通过水解β-1, 4糖苷键从而破坏木聚糖的主链。木聚糖酶作为饲料酶可以有效解除木聚糖引起的抗营养作用,用于淀粉-谷朊粉分离以提高淀粉的得率,因此木聚糖酶在饲料[1]、食品加工[2]和纸浆漂白[3]等领域具有重要的作用。
小麦中共有3种类型的木聚糖酶抑制蛋白,分别为Triticum aestivum xylanase inhibitor (TAXI)[4]、Xylanase inhibitor protein (XIP)[5]、Thaumatin-like xylanase inhibitor (TLXI)[6]。Croes等[7]利用免疫印迹的方法对8个欧洲小麦品种的木聚糖酶抑制蛋白进行测定,发现以XIP型的抑制蛋白含量最高,平均含量达到235 μg/kg。因此在添加木聚糖酶的谷物加工领域中,抑制蛋白的广泛且大量存在对木聚糖酶的用酶效果产生了不利影响。鉴于此,研究XIP型抑制蛋白与木聚糖酶相互作用的分子机制,获得XIP型木聚糖酶抑制蛋白的抗性木聚糖酶将最大程度地发挥外加木聚糖酶的应用效果。
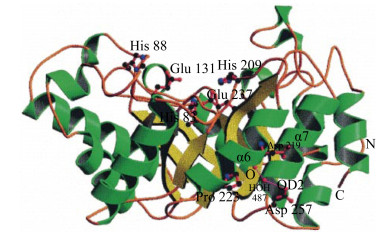
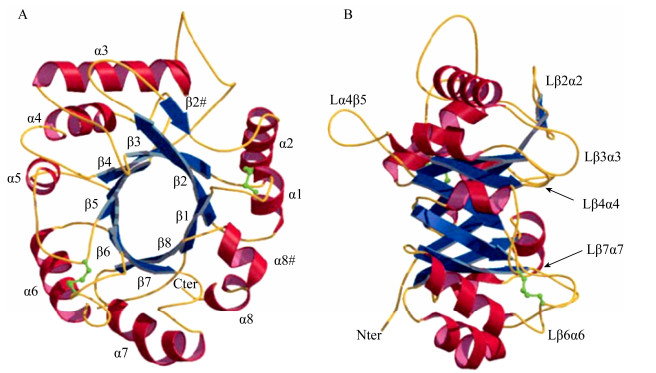
2 GH10和GH11木聚糖酶的分子结构不同来源木聚糖酶的氨基酸数目相差很大,但催化区域的大小比较一致。根据催化区域保守氨基酸序列和疏水簇的分析,木聚糖酶主要集中在糖苷水解酶GH10家族和GH11家族,同一家族的木聚糖酶催化区域具有较高的相似性[8]。GH10家族木聚糖酶是由(α/β)8构成的桶状结构,形似“色拉碗”,底物结合缝隙位于桶状结构内部β折叠的C末端,催化中心是两个高度保守的Glu (图 1)[9]。GH10家族木聚糖酶一般拥有4–6个底物结合亚位点,其中–2、–1、+1亚位点高度保守并展现出与底物的高亲和力,导致GH10家族木聚糖酶能够催化木聚糖产生低聚合度的木寡糖(图 2)[10]。
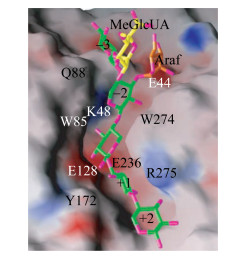
GH11家族木聚糖酶由一个α螺旋与两个反向平行的β折叠片构成,形似“半握状的右手”,拥有一个简单的催化结构域,催化中心两个保守的Glu位于“手掌”状的凹面底物结合缝隙中,亚位点的数目一般为5–6个,并且水解反应发生在–1、+1亚位点之间;位于底物结合缝隙上方的两个β折叠股之间的Loop被称作“拇指” (thumb)结构,GH11家族木聚糖酶中“拇指”结构的长短各不相同,其通过疏水作用力和氢键保持自身的稳定,较高的灵活性可以使其自由移动来调整底物结合缝隙入口的宽度(图 3)[11]。
通过X-ray得知XIP型木聚糖酶抑制蛋白的三维结构是一个由(β/α)8构成的椭圆骨架,其顶端的环沿着分子的一面形成一个凹陷;在XIP型木聚糖酶抑制蛋白的活性中心存在3个保守的芳香族氨基酸残基,分别是Trp256XIP-I、Tyr189XIP-I和Phe11XIP-I;XIP型抑制蛋白通过底物模拟方式竞争性抑制木聚糖酶(图 4)[12]。XIP型的抑制蛋白能够同时抑制来自真菌大部分GH10和GH11木聚糖酶的活力,但对不同结构木聚糖酶的抑制常数Ki变化范围较大[13-14]。
目前,XIP型木聚糖酶抑制蛋白对GH10家族真菌木聚糖酶的抑制机制尚不十分清楚。已经解析的XIP-I蛋白与Aspergillus nidulans木聚糖酶XLNC复合体的晶体结构显示(图 5A),XIP-I与XLNC活性位点的结合致使木聚糖酶的底物结合缝隙被完全覆盖,底物分子无法正常进出酶的底物结合缝隙;二者相互作用的面积达到了1 130Å2,位于XIP-I α7 (232–245)肽段中的Lys234XIP-I、Asn235XIP-I、His232XIP-I和Tyr238XIP-I等4个氨基酸残基牢牢地占据了XLNC的–1、+1、+2和−2亚位点,同时Lys246XIP-I阻塞了底物接近−3亚位点的通道;各亚位点区域关键的底物结合位点Trp269XLNC、His209XLNC、Trp277XLNC、Tyr174XLNC、Arg278XLNC分别与Tyr237XIP-I、Tyr238XIP-I、His232XIP-I、Trp230XIP-I、Tyr273XIP-I相结合,从而阻碍底物分子的结合;Lys234XIP-I则直接与的酸碱催化残基Glu131XLNC形成氢键相连,同时通过与水分子形成的氢键间接地与起亲核攻击作用的Glu239XLNC相连。酶的催化中心被XIP-I以氢键的形式牢牢结合,彻底失去了水解糖苷键的机会[15]。
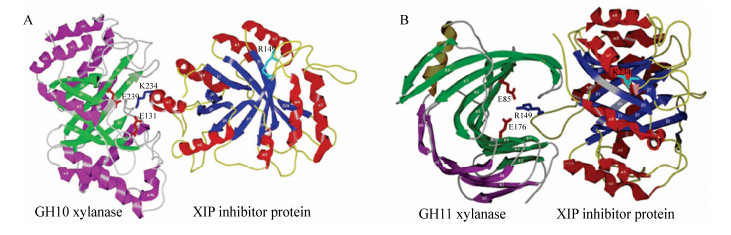
|
| 图 5 XIP型木聚糖酶抑制蛋白与GH10、GH11木聚糖酶的相互作用模式图[15] Figure 5 Interaction pattern of XIP-type xylanase inhibitor protein with GH10 and GH11 xylanase[15] 注:A:XIP型木聚糖酶抑制蛋白与GH10木聚糖酶;B:XIP型木聚糖酶抑制蛋白与GH11木聚糖酶. Note: A: XIP-type xylanase inhibitor protein and GH10 xylanase; B: XIP-type xylanase inhibitor protein and GH11 xylanase. |
|
|
XIP型木聚糖酶抑制蛋白对GH11家族木聚糖酶的抑制机理比较清楚。Penicillium funiculosum木聚糖酶XYNC和XIP-I抑制蛋白复合体的晶体结构显示(图 5B),XIP-I蛋白的α4与β5之间的Π型Loop (148–154)和木聚糖酶XYNC的活性中心、“拇指”结构、“手掌”结构区域同时作用,占据了酶的活性位点并堵塞了底物进入活性位点区域的通道;该Loop的顶端轻微地扭曲,导致处于150–152位的3个氨基酸Gly-Gly-Pro在Gly150XIP-I处转向平行延伸至–3亚位点,阻碍底物接近,同时XIP蛋白的Gly150XIP-I的主链羰基和木聚糖酶XYNC上被预测为可以与木糖环结合的Ser16XYNC相互作用;Arg149XIP-I、Gly150XIP-I的主链片段则牢牢占据了–2亚位点,Arg149XIP-I的侧链从Loop的左侧边缘展露出来,与高度保守的Trp18XYNC形成疏水作用力,模拟木糖环在–2亚位点堆叠;同时,Arg149XIP-I的胍基进入–1亚位点,与酸碱催化碱基Glu176XYNC和Tyr87XYNC以氢键的形式相结合,阻止了降解产物的释放[15-16]。此外,XIP-I的α4β5、α5β6和α6β7之间的Loop与XYNC的“拇指”结构也有着紧密的结合[15]。最近研究者通过分子动力学模拟揭示了水稻木聚糖酶抑制(rice xylanase inhibitor,RIXI)蛋白中一个长的Loop结构(144–153位点)插入GH11木聚糖酶的催化裂隙中;非变性PAGE检测到XIP型木聚糖酶抑制蛋白与木聚糖酶相互作用后形成了复合物;同时在活酵母细胞中也观察到了RIXI蛋白和木聚糖酶之间相互作用[17]。
5 对XIP型木聚糖酶抑制蛋白具有抗性的木聚糖酶研究表明,XIP型木聚糖酶抑制蛋白对隶属于GH10家族的植物木聚糖酶以及细菌来源的GH10和GH11木聚糖酶并未展现出抑制性[13],因此上述三类木聚糖酶均为XIP抗性木聚糖酶,另外个别真菌来源的野生型木聚糖酶也对XIP型木聚糖酶抑制蛋白表现出明显的抗性[18-21]。
5.1 细菌和真菌来源的GH10木聚糖酶Pseudomonas fluorescens木聚糖酶Xyn10A不受XIP-I抑制蛋白所抑制;序列比对显示,在与XIP-I相互作用区域,Xyn10A与XLNC两个不同来源GH10木聚糖酶的氨基酸序列之间并无明显的区别,然而木聚糖酶Xyn10A中连接α4和β4的Loop以及连接α8和β8的Loop上插入了更多的氨基酸,从而导致Loop明显增长;据推测,这些长的Loop在空间上排斥XIP-I接近酶的底物结合缝隙;这种较长Loop结构同样存在于唯一不受XIP-I所抑制的Aspergillus aculeatus GH10木聚糖酶中,在其β7和β8后同样插入了较多的氨基酸[13-14]。
在生物质降解过程中,GH10家族木聚糖酶比GH11家族木聚糖酶具有更好的催化性能,因此在酶的应用中更具有经济可行性[22],然而关于真菌GH10木聚糖酶XIP抗性改造的研究报道较少,目前在PubMed、ScienceDirect和Web of Science数据库中只查到2019年的一篇文献,该文献中Denisenko等[23]将真菌来源的2种XIP敏感性木聚糖酶与4种XIP抗性木聚糖酶进行了序列比对及空间结构叠加,确定了靠近抗性木聚糖酶活性位点的Loop环插入(含GQGGA肽段)可能是影响XIP抑制作用的关键因素;他们进一步对来自于P. canescens的XIP敏感性GH10家族木聚糖酶PcXylA和T. reesei来源的GH10家族木聚糖酶TrXyn3进行了蛋白质工程改造,在活性位点裂口的Loop环中插入GQGGA 5个氨基酸残基,结果表明该Loop环的延长导致木聚糖酶对黑麦提取物中的蛋白质抑制剂产生了抗性。
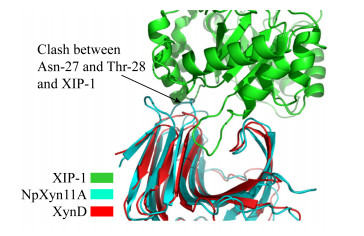
5.2 真菌来源的GH11木聚糖酶动物瘤胃厌氧真菌新丽鞭毛菌(Neocallimastix patriciarum)来源的GH11木聚糖酶NpXyn11A对XIP-I抑制蛋白具有天然的强烈抗性;相比其他GH11木聚糖酶,NpXyn11A的“拇指”结构区域(氨基酸序列为TGPTINGGSE)顶端插入了Gly154NpXyn11A,导致“拇指”结构比其他GH11家族木聚糖酶更加凸出,并且Gly154NpXyn11A的氨基和α碳原子与Ala144XIP-I的羰基团相互排斥;另一方面,NpXyn11A中位于β3与β4 Loop上的Leu25-Asn31均与XIP-I在空间上形成对抗;此外,NpXyn11A宽广的底物结合缝隙也减少了XIP-I的Π型Loop与酶结合位点相互作用的频率,甚至无法接触结合位点(图 6)[18]。我们将Neocallimastix sp. GMLF1的XIP抗性木聚糖酶Xyn1B (GenBank登录号:ACG68418.1)与A. niger的XIP敏感木聚糖酶(UniProt ID:P55328)及P. funiculosum的XIP敏感木聚糖酶XYNC (UniProt:Q9HFH0)进行序列比对,发现XIP抗性木聚糖酶的“拇指”部位氨基酸发生了GTS到GGSE的突变[24]。然而将抗性木聚糖酶Xyn1B的“拇指”结构162GPTINGGSETF172中的167GGSE170突变为GTS,甚至将Xyn1B的“拇指”序列完全由P. funiculosum木聚糖酶XYNC的“拇指”序列来替换,得到的突变酶对XIP的抗性并没有发生变化;依据XIP-I与抗性木聚糖酶Xyn1B以及P. funiculosum敏感性木聚糖酶XYNC相互作用的结构模型,将Xyn1B的“手掌”结构区域中β3-β4环及β4-β5环同时突变为木聚糖酶XYNC的相应序列,以扩大酶的底物结合缝隙,然而所得突变酶的XIP抗性仍未受到影响;由此推测瘤胃真菌的抗性木聚糖酶Xyn1B可能具有独特的分子结构,其对XIP型木聚糖酶抑制蛋白保持抗性并不仅仅是某个氨基酸或某个多肽片段的影响[25-26]。
植物病原真菌Fusarium graminearum来源的GH11木聚糖酶XylA和XylB经原核表达得到的重组酶,同样不能被XIP-I抑制[19],然而XIP-I抑制能力损失与糖基化无关[27]。通过与其他GH11家族木聚糖酶序列比对,发现Aspergillus niger木聚糖酶“拇指”结构中高度保守的Gly后面的Thr被木聚糖酶XylA中Val151所替代,而Val151及其侧链在空间上与XIP-I相斥,是木聚糖酶XlyA获得抗性的原因;然而Thr侧链可以与XIP-I的Asn147形成氢键,从而使酶受到XIP-I的抑制;木聚糖酶XylB的“拇指”结构中两个特有的Cys存在导致XylB无法与XIP-I的Gly179、Ala214、Asn147之间形成氢键结合,从而产生抗性(图 7)[28]。
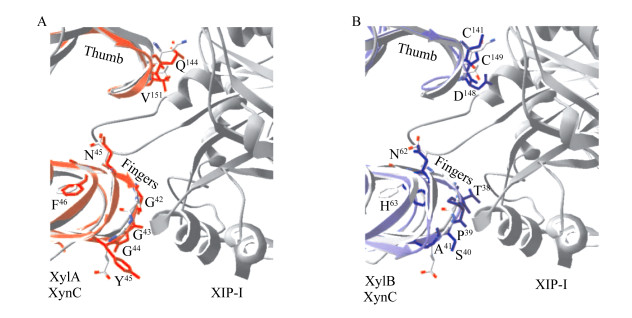
|
| 图 7 叠合有木聚糖酶XynC的F. graminearum GH11木聚糖酶XylA和XylB与XIP-I的复合物结构[28] Figure 7 The complex structure of the XIP-I/F. graminearum GH11 xylanases XylA and XylB overlaid with P. funiculosum XynC[28] 注:A:木聚糖酶XylA与XIP-I抑制蛋白;B:木聚糖酶XylB与XIP-I抑制蛋白. Note: A: Xylanase XylA and XIP-I inhibitor protein; B: Xylanase XylB and XIP-I inhibitor protein. |
|
|
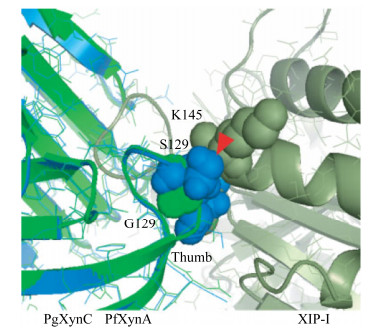
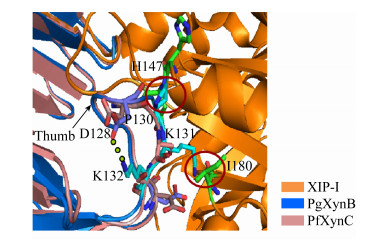
相较于上述XIP抗性GH11木聚糖酶,Penicillium griseofulvum木聚糖酶PgXynA的“拇指”结构区域(氨基酸序列为NQPSIISDSS)在相同的位置同样有一个额外的氨基酸Asp130的插入,并且高度保守的Gly被Ser129替换;Ser129、Asp130组合突变酶的抗抑制性损失严重,Ki值变成了3.9×10−12 mol/L;模型分析显示,Ser129、Asp130的侧链分别与XIP-I的Lys145、Asn147形成对抗(图 8)[20]。在Penicillium griseofulvum来源的另一种木聚糖酶PgXynB中,将其“拇指”结构区域(氨基酸序列为DQPSIDGPKK)插入的Pro130删除,同时将随后的两个Lys突变成Ser,突变体酶失去了对XIP-I的抗性,抑制常数Ki变为16.5×10−12 mol/L;根据PgXynB与XIP型木聚糖酶抑制蛋白复合体的模型分析,木聚糖酶PgXynB的Pro130和Lys131分别与XIP-I的Asn146、His147和Ile180在空间上形成了对抗,并且木聚糖酶PgXynB的Lys132与Asp128之间形成的氢键使得“拇指”结构韧性更强,从而无法灵活地调节进入底物结合缝隙的开口大小,使其对XIP型木聚糖酶抑制蛋白产生了抗性(图 9)[21]。因此,“拇指”结构灵活性的降低在抗XIP-I的过程中起着至关重要的作用,而木聚糖酶PgXynA、PgXynB对XIP蛋白的抗性并不完全是由于“拇指”结构区域单个氨基酸的插入所导致。
XIP-I对Aspergillus niger的GH11木聚糖酶具有强烈的抑制性,Tahir等[29]通过单点突变构建了部分突变体,当第117位Asn突变成Ala后,对XIP-I敏感的Aspergillus niger木聚糖酶获得了抗性;Asn117位于β8的末端,毗邻“拇指”结构,并且高度保守;结构复杂的Asn突变为简单的Ala后致使“拇指”结构滑动,导致突变体酶的底物结合缝隙变窄,从而阻止了大分子量的XIP型木聚糖酶抑制蛋白进入;同时,“拇指”结构区域的一些氨基酸Thr114、Thr116、Asn117、Thr124、Thr126和Thr128,包括其侧链基团均与XIP-I形成空间上的对抗。
6 结论与展望XIP-I与木聚糖酶的相互作用过程十分迅速,通过模拟底物接触方式竞争性地结合酶的活性位点,阻止底物进出底物结合缝隙。细菌GH10木聚糖酶产生的XIP抗性,可能是部分连接α/β之间的Loop结构较长,因而造成连接各α/β的Loop长短不一,从而在较远的距离就能与XIP-I接触,导致XIP-I无法接近底物结合缝隙。
XIP作用于GH11家族真菌木聚糖酶主要是XIP的Π型Loop堵塞了进入活性位点区域的通道。在大部分XIP抗性GH11木聚糖酶中,“拇指”结构区域有额外的氨基酸插入,同时伴有保守氨基酸被替换,导致“拇指”构象变化以及灵活性的降低,不能调节进入底物结合缝隙入口的宽度。
外源木聚糖酶抑制蛋白对不同的微生物木聚糖酶有着复杂的识别特异性,这种识别特异性是植物为防御来自不同病原菌的多种木聚糖酶侵害而不断进化的结果。XIP型外源木聚糖酶抑制蛋白的存在贯穿整个植物的生命阶段,在植物的生长过程以及防御致病真菌的入侵过程中起着重要的作用[30],同时通过过量表达实验还发现水稻XIP型木聚糖酶抑制剂Oshi-XIP参与了水稻对危害最为严重的常发性害虫二化螟的抗性[31],因此,构建XIP过量表达的植株将有效提高其抗病抗虫能力。另一方面,利用厌氧真菌Neocallimastix sp.产生的XIP天然抗性木聚糖酶用于麦芽汁糖化,可以有效抵抗外加XIP型木聚糖酶抑制蛋白的不利作用,提高木聚糖酶的用酶效果[19]。Frederix等[32]在谷朊粉-淀粉的分离过程中外加对TAXI及XIP型木聚糖酶抑制蛋白同时具有抗性的两种木聚糖酶,发现棘孢曲霉木聚糖酶XAA具有显著的改善效果,外加枯草杆菌木聚糖酶XBSni时也只需要较少的酶量即可提高分离效果。因此依据抑制蛋白与木聚糖酶的相互作用机制对商业常用真菌木聚糖酶进行蛋白质工程改造,获得抑制蛋白抗性的高活性木聚糖酶突变体用于食品加工将有效提高用酶效果。
| [1] |
He X, Yu B, He J, et al. Effects of xylanase on growth performance, nutrients digestibility and intestinal health in weaned piglets[J]. Livestock Science, 2020, 233: 103940. DOI:10.1016/j.livsci.2020.103940 |
| [2] |
Adiguzel G, Faiz O, Sisecioglu M, et al. A novel endo-β-1, 4-xylanase from Pediococcus acidilactici GC25; purification, characterization and application in clarification of fruit juices[J]. International Journal of Biological Macromolecules, 2019, 129: 571-578. DOI:10.1016/j.ijbiomac.2019.02.054 |
| [3] |
Campioni TS, de Jesus Moreira L, Moretto E, et al. Biobleaching of Kraft pulp using fungal xylanases produced from sugarcane straw and the subsequent decrease of chlorine consumption[J]. Biomass and Bioenergy, 2019, 121: 22-27. DOI:10.1016/j.biombioe.2018.12.014 |
| [4] |
Debyser W, Peumans WJ, van Damme EJM, et al. Triticum aestivum xylanase inhibitor (TAXI), a new class of enzyme inhibitor affecting breadmaking performance[J]. Journal of Cereal Science, 1999, 30(1): 39-43. |
| [5] |
McLauchlan WR, Garcia-Conesa MT, Williamson G, et al. A novel class of protein from wheat which inhibits xylanases[J]. Biochemical Journal, 1999, 338(2): 441-446. |
| [6] |
Fierens E, Rombouts S, Gebruers K, et al. TLXI, a novel type of xylanase inhibitor from wheat (Triticum aestivum) belonging to the thaumatin family[J]. Biochemical Journal, 2007, 403(3): 583-591. |
| [7] |
Croes E, Gebruers K, Luyten N, et al. Immunoblot quantification of three classes of proteinaceous xylanase inhibitors in different wheat (Triticum aestivum) cultivars and milling fractions[J]. Journal of Agricultural and Food Chemistry, 2009, 57(3): 1029-1035. DOI:10.1021/jf802638n |
| [8] |
Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases[J]. FEMS Microbiology Reviews, 2005, 29(1): 3-23. DOI:10.1016/j.femsre.2004.06.005 |
| [9] |
Natesh R, Bhanumoorthy P, Vithayathil PJ, et al. Crystal structure at 1.8Å resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus[J]. Journal of Molecular Biology, 1999, 288(5): 999-1012. |
| [10] |
Fujimoto Z, Kaneko S, Kuno A, et al. Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86[J]. Journal of Biological Chemistry, 2004, 279(10): 9606-9614. DOI:10.1074/jbc.M312293200 |
| [11] |
Paës G, Berrin JG, Beaugrand J. GH11 xylanases: structure/function/properties relationships and applications[J]. Biotechnology Advances, 2012, 30(3): 564-592. DOI:10.1016/j.biotechadv.2011.10.003 |
| [12] |
Payan F, Flatman R, Porciero S, et al. Structural analysis of xylanase inhibitor protein I (XIP-I), a proteinaceous xylanase inhibitor from wheat (Triticum aestivum, var. Soisson)[J]. Biochemical Journal, 2003, 372(2): 399-405. |
| [13] |
Flatman R, McLauchlan WR, Juge N, et al. Interactions defining the specificity between fungal xylanases and the xylanase-inhibiting protein XIP-I from wheat[J]. Biochemical Journal, 2002, 365(3): 773-781. |
| [14] |
Goesaert H, Elliott G, Kroon PA, et al. Occurrence of proteinaceous endoxylanase inhibitors in cereals[J]. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2004, 1696(2): 193-202. DOI:10.1016/j.bbapap.2003.08.015 |
| [15] |
Payan F, Leone P, Porciero S, et al. The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases[J]. The Journal of Biological Chemistry, 2004, 279(34): 36029-36037. DOI:10.1074/jbc.M404225200 |
| [16] |
Sidhu G, Withers SG, Nguyen NT, et al. Sugar ring distortion in the glycosyl-enzyme intermediate of a family G/11 xylanase[J]. Biochemistry, 1999, 38(17): 5346-5354. DOI:10.1021/bi982946f |
| [17] |
Liu MQ, Wu XQ, Huo WK, et al. Differential inhibition of GH family 11 endo-xylanase by rice xylanase inhibitor and verification by a modified yeast two-hybrid system[J]. International Journal of Biological Macromolecules, 2019, 132: 514-523. DOI:10.1016/j.ijbiomac.2019.04.001 |
| [18] |
Vardakou M, Dumon C, Murray JW, et al. Understanding the structural basis for substrate and inhibitor recognition in eukaryotic GH11 xylanases[J]. Journal of Molecular Biology, 2008, 375(5): 1293-1305. |
| [19] |
Beliën T, van Campenhout S, van Acker M, et al. Cloning and characterization of two endoxylanases from the cereal phytopathogen Fusarium graminearum and their inhibition profile against endoxylanase inhibitors from wheat[J]. Biochemical and Biophysical Research Communications, 2005, 327(2): 407-414. DOI:10.1016/j.bbrc.2004.12.036 |
| [20] |
André-Leroux G, Berrin JG, Georis J, et al. Structure-based mutagenesis of Penicillium griseofulvum xylanase using computational design[J]. Proteins, 2008, 72(4): 1298-1307. |
| [21] |
Tison MC, André-Leroux G, Lafond M, et al. Molecular determinants of substrate and inhibitor specificities of the Penicillium griseofulvum family 11 xylanases[J]. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2009, 1794(3): 438-445. DOI:10.1016/j.bbapap.2008.11.024 |
| [22] |
Hu JG, Saddler JN. Why does GH10 xylanase have better performance than GH11 xylanase for the deconstruction of pretreated biomass?[J]. Biomass and Bioenergy, 2018, 110: 13-16. DOI:10.1016/j.biombioe.2018.01.007 |
| [23] |
Denisenko YA, Gusakov AV, Rozhkova AM, et al. Protein engineering of GH10 family xylanases for gaining a resistance to cereal proteinaceous inhibitors[J]. Biocatalysis and Agricultural Biotechnology, 2019, 17: 690-695. DOI:10.1016/j.bcab.2019.01.042 |
| [24] |
Liu XY. Properties of xylanase inhibitor protein from wheat and its influence on efficacy of exogenous xylanases[D]. Zhengzhou: Doctoral Dissertation of Henan Agricultural University, 2016 (in Chinese) 刘新育.小麦木聚糖酶抑制蛋白的性质及对外源木聚糖酶功效的影响研究[D].郑州: 河南农业大学博士学位论文, 2016 |
| [25] |
Zhu DD. Structure elements for inhibitor protein resistance of Xylanse Xyn1B and its application experiment[D]. Zhengzhou: Master's Thesis of Henan Agricultural University, 2018 (in Chinese) 朱东东.抗抑制性木聚糖酶Xyn1B抗性结构基础及抗性酶应用试验[D].郑州: 河南农业大学硕士学位论文, 2018 |
| [26] |
Zhu DD, Liu XY, Xie X, et al. Characteristics of a XIP-resistant xylanase from Neocallimastix sp. GMLF1 and its advantage in barley malt saccharification[J]. International Journal of Food Science & Technology, 2020, 55(5): 2152-2160. |
| [27] |
Juge N, Payan F, Williamson G. XIP-I, a xylanase inhibitor protein from wheat: a novel protein function[J]. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 2004, 1696(2): 203-211. DOI:10.1016/j.bbapap.2003.08.014 |
| [28] |
Beliën T, van Campenhout S, van Acker M, et al. Mutational analysis of endoxylanases XylA and XylB from the phytopathogen Fusarium graminearum reveals comprehensive insights into their inhibitor insensitivity[J]. Applied and Environmental Microbiology, 2007, 73(14): 4602-4608. DOI:10.1128/AEM.00442-07 |
| [29] |
Tahir TA, Berrin JG, Flatman R, et al. Specific characterization of substrate and inhibitor binding sites of a glycosyl hydrolase family 11 xylanase from Aspergillus niger[J]. Journal of Biological Chemistry, 2002, 277(46): 44035-44043. DOI:10.1074/jbc.M205657200 |
| [30] |
Sun RJ, Xu Y, Hou CX, et al. Expression and characteristics of rice xylanase inhibitor OsXIP, a member of a new class of antifungal proteins[J]. Biologia Plantarum, 2018, 62(3): 569-578. DOI:10.1007/s10535-018-0787-2 |
| [31] |
Xin ZJ, Wang Q, Yu ZN, et al. Overexpression of a xylanase inhibitor gene, OsHI-XIP, enhances resistance in rice to herbivores[J]. Plant Molecular Biology Reporter, 2014, 32(2): 465-475. DOI:10.1007/s11105-013-0661-5 |
| [32] |
Frederix SA, Courtin CM, Delcour JA. Substrate selectivity and inhibitor sensitivity affect xylanase functionality in wheat flour gluten–starch separation[J]. Journal of Cereal Science, 2004, 40(1): 41-49. DOI:10.1016/j.jcs.2004.04.002 |
 2020, Vol. 47
2020, Vol. 47