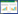扩展功能
文章信息
- 吴秀芸, 戴琳, 张舒, 尚伟昊, 余俊红, 黄小平, 王禄山
- WU Xiu-Yun, DAI Lin, ZHANG Shu, SHANG Wei-Hao, YU Jun-Hong, HUANG Xiao-Ping, WANG Lu-Shan
- 高效木聚糖降解酶系定制的新进展与挑战
- New advances and challenges in customizing xylan-degrading enzymes
- 微生物学通报, 2020, 47(7): 2278-2289
- Microbiology China, 2020, 47(7): 2278-2289
- DOI: 10.13344/j.microbiol.china.200316
-
文章历史
- 收稿日期: 2020-03-30
- 网络首发日期: 2020-06-12
2. 青岛啤酒股份有限公司啤酒生物发酵工程国家重点实验室 山东 青岛 266000;
3. 内蒙古伊利实业集团股份有限公司创新中心 内蒙古 呼和浩特 010110
2. State Key Laboratory of Biological Fermentation Engineering of Beer, Tsingtao Brewery Company Limited, Qingdao, Shandong 266000, China;
3. Innovation Center of Inner Mongolia Yili Industrial Group Company Limited, Hohhot, Inner Mongolia 010110, China
生物炼制工业以低价值的木质纤维素为原材料,酸解、碱解及化学试剂的使用既要耗费大量淡水,又造成环境污染。酶解作为一种绿色生产工艺,不仅可以减少水的利用和能源消耗,而且其作用专一、转化效率高。但是木质纤维素的物理结构复杂且具有异质性,特别是木聚糖类的底物更加复杂多样,不同来源木聚糖的物理化学性质各不相同,因此,必须针对特定类型的木聚糖优化生物炼制工艺。只有了解不同底物富含木聚糖的主要结构特点和木聚糖降解酶的底物特异性,才能针对不同的底物选择偏好降解的酶系或微生物,进而通过优化原料组合和配比达到高效降解的目标。将底物复杂性与木聚糖降解酶组分精准配制,设计改造并优化组合来定制木聚糖降解酶系,从而实现对木质纤维素资源的绿色高值化利用。
1 木聚糖结构的异质性木聚糖是自然界中第二丰富的可再生多糖,由木糖单体组成主链,一般具有不同取代基的侧链(表 1)。直链木聚糖(β-1, 3和/或β-1, 4糖苷键连接)只有主链结构,无侧链取代基,主要存在于低等植物中,如绿藻[1]、红藻[2]和毛瑞榈[6]。异质性木聚糖不仅具有β-1, 4-连接的木聚糖主链,还具有各种糖单元为取代基的侧链,取代基可以是葡萄糖醛酸、阿拉伯糖、木糖、半乳糖和葡萄糖等,不同植物来源或不同植物组织中的木聚糖,其侧链的取代基和取代度不尽相同[3, 7],分别命名为葡萄糖醛酸木聚糖、阿拉伯木聚糖、葡萄糖醛酸阿拉伯木聚糖(图 1)。
| 木聚糖类型 Xylan |
植物来源 Sources |
主骨架连接键 Back-bone linkage |
主要侧链取代基 Substituent group |
参考文献 References |
| 直链木聚糖 Linear-xylan |
海藻 Seaweeds |
β-1, 4, β-1, 3 | 甲基 Methyl-groups |
[1-2] |
| 葡萄糖醛酸木聚糖 Glucoronoxylan |
木本植物 Xylophyta |
β-1, 4 | 乙酰基、葡萄糖醛酸 Acetyl-groups, glucuronic acid |
[3] |
| 阿拉伯木聚糖 Arabinoxylan |
草本植物 Grasses |
β-1, 4 | 乙酰基、阿拉伯糖 Acetyl-groups, arabinose |
[4] |
| 葡萄糖醛酸阿拉伯木聚糖 Glucuronoarabinoxylan |
玉米等单子叶植物 Monocotyledon (corn) |
β-1, 4 | 乙酰基、葡萄糖醛酸、阿拉伯糖 Acetyl-groups, glucuronic acid, arabinose |
[3, 5] |
葡萄糖醛酸木聚糖(glucoronoxylan,GX)是木本植物中主要的半纤维素多糖[3]。阔叶木材中的木聚糖是O-乙酰基-4-O-甲基葡萄糖醛酸-β-D-木聚糖(有时也称为乙酰葡萄糖醛酸木聚糖),占木材干重的15%−35%[9]。乙酰基在木聚糖链上有规律的分布,几乎每隔一个木糖就有O-2或O-3单取代(34%−49%)或双取代(6%−7%)的乙酰基[10-11],当O-2被4-O-甲基葡萄糖醛酸取代时,O-3可同时被乙酰基取代(10%)[12],木聚糖的聚合度(degree of polymerization,DP)大约为200;针叶木材中的木聚糖含量较低,而DP通常较低,大约为100[13]。糖醛酸在局部的分布是有规律的,每10个木糖残基内O-2位有2个4-O-甲基葡萄糖醛酸[14]。
1.2 阿拉伯木聚糖(arabinoxylan,AX)阿拉伯木聚糖(arabinoxylan,AX)是草本植物中主要的半纤维素多糖[4]。阿拉伯木聚糖中侧链取代基为阿拉伯糖,它们一般连接在主链结构中木糖单元的O-2和/或O-3位;阿拉伯木聚糖的平均DP在50−185之间,乙酰化程度低于葡萄糖醛酸木聚糖[15]。另外,阿拉伯糖侧基C-5位可能有阿魏酸取代基的存在,阿拉伯木聚糖可以通过这些阿魏酸基团与木质素共价交联[16]。但是,由于阿魏酸是酚基,因此应将其视为木质素而非半纤维素的构建基块。
1.3 葡萄糖醛酸阿拉伯木聚糖(glucuronoarabinoxylan,GAX)单子叶植物次生细胞壁木聚糖中,侧链取代基同时含有4-O-甲基葡萄糖醛酸(和/或葡萄糖醛酸)基团和阿拉伯糖基团,阿拉伯糖存在单取代或双取代,在O-2或O-3位,O-3位阿拉伯糖的O-2位进一步可被阿拉伯糖或木糖苷取代,O-5位可被阿魏酸或p-香豆酸酯化[17]。另外,复杂木聚糖结构中木聚糖主链与寡糖侧链相连,该寡糖侧链由各种官能团组成,例如4-O-甲基葡萄糖醛酸、葡萄糖醛酸、阿拉伯糖、木糖、半乳糖、葡萄糖和岩藻糖等[3, 5]。
通过对不同底物的单糖组成单元分析(表 2)发现,来源于单子叶植物(小麦麸皮、玉米皮)和双子叶植物(豆渣、豆皮)的粮食加工副产物的化学组成和结构层次具有明显的差异[18]。单子叶植物的主要成分是纤维素和木聚糖,而双子叶植物中主要是果胶、甘露聚糖及木葡聚糖成分。单子叶植物小麦麸皮和玉米皮所含单糖种类相同,但是含量存在差异[18]。研究表明,微生物产木聚糖酶不能彻底降解小麦阿拉伯木聚糖(wheat arabinoxylan,WAX;Ara:Xyl=39:61,摩尔比),在反应体系中加入侧链降解酶阿拉伯呋喃糖苷酶会加速木聚糖的降解,提高单糖的产量[19]。相比于小麦麸皮,玉米皮的复杂糖结构被不同酯化基团取代如阿魏酸、香豆酸等,其高效降解还需要阿魏酸酯酶及香豆酸酯酶的添加,因此需要针对不同底物(材料)富含木聚糖的主要成分的结构特点来设计酶系。
| 样品 Sample |
葡萄糖 Glucose |
木糖 Xylose |
阿拉伯糖 Arabinose |
半乳糖 Galactose |
甘露糖 Mannose |
鼠李糖 Rhamnose |
岩藻糖 Fucose |
| 小麦麸皮 Wheat bran |
40.5 | 22.4 | 30.8 | 5.3 | 1.0 | − | − |
| 玉米皮 Corn bran |
33.1 | 29.1 | 34.3 | 2.9 | 0.6 | − | − |
| 豆渣 Soybean dregs |
7.4 | 11.9 | 6.2 | 42.6 | 1.6 | 27.9 | 2.5 |
| 豆皮 Soybean hulls |
7.9 | 23.9 | 7.3 | 14.2 | 23.0 | 21.6 | 2.1 |
| Note: “−” Nothing was detected. | |||||||
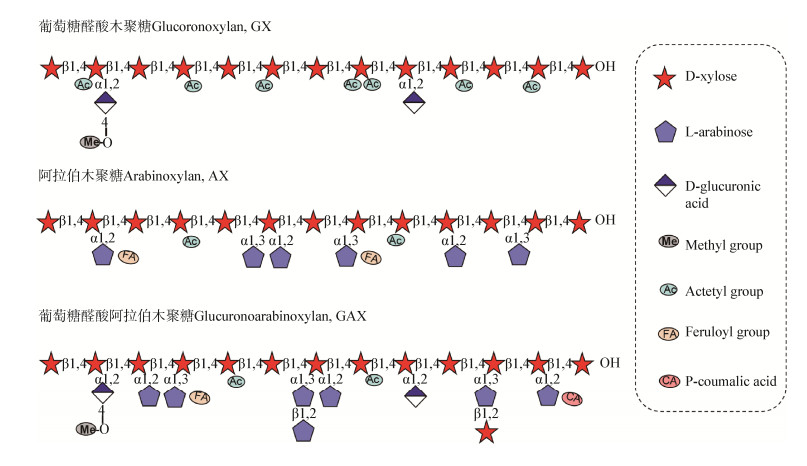
木聚糖的复杂结构与基因组中参与细胞壁合成基因的多样性和差异性表达息息相关[20]。木聚糖合成酶系包括主链合成酶系和侧基转移酶系,这些酶类大多位于高尔基体中,因此木聚糖的合成是在高尔基体上完成并通过囊泡运输将多糖分泌到胞外[17]。近年来,植物木聚糖主链合成和取代基合成取得了较大的研究进展(图 2)。
2.1 主链合成酶细胞质中的UDP-木糖合成酶(UDP-xylose synthase,UXS)催化UDP-葡萄糖醛酸形成UDP-木糖,然后由UDP-木糖转运体转运至高尔基体内腔,位于高尔基体的糖苷转移酶GT43家族IRX9、IRX14和GT47家族的IRX10木聚糖合成酶复合体(xylan synthase complex)催化合成木聚糖主链骨架[17, 21]。突变木聚糖合成酶复合体中任意一个组分都会造成植物矮化以及木聚糖含量和糖基转移酶活性的降低[22-23]。此外,Peña等对于木聚糖主链骨架合成提出了另一个模型,即短链(1, 4)-β-D-木聚糖的合成是由木四糖启动的,GT8家族的IRX8、PARVUS以及GT47家族的IRX7/FRA8编码的木聚糖合成酶催化合成初始木四糖[24]。GT8家族编码保留型转移酶,催化α-D-糖苷键的形成;GT47家族编码转化型转移酶,催化β-D-糖苷键或α-L-糖苷键的形成[25]。
2.2 侧基转移酶葡萄糖醛酸取代基是由GT8家族的木聚糖葡萄糖醛酸转移酶(xylan α-glucuronyltransferase,GUX)家族催化,其中在GUX1和GUX2的作用下引入木聚糖主链骨架上[26],另外,GT8家族还包括GUX3、GUX4和GUX5。GT8家族的木聚糖葡萄糖醛酸基转移酶是将未甲基化葡萄糖醛酸添加到木聚糖骨架上[27-28],之后,葡萄糖醛酸的4-OH位置被葡萄糖醛酸木聚糖甲基转移酶(glucuronate xylan methyltransferase,GXMT)进行甲基化[17]。阿拉伯木聚糖合成过程与葡萄糖醛酸木聚糖不同,UDP-木糖在高尔基体中可被UDP-木糖差向异构酶(UDP-xylose epimerase,UXE)催化形成UDP-阿拉伯吡喃糖,UDP-阿拉伯吡喃糖转化为UDP-阿拉伯呋喃糖是由位于高尔基体外膜上的可逆糖化蛋白(reversibly glycosylated protein,RGP)所催化,然后,UDP-阿拉伯呋喃糖被转运蛋白转运到高尔基体内部,由GT61家族阿拉伯呋喃糖苷转移酶(xylan arabinosyltransferase,XAT)在主链上添加α-L-阿拉伯呋喃糖苷[29-30]。异质性木聚糖的乙酰基在主链骨架上是规律分布的,主要连接在主链木糖的2-OH和/或3-OH,每隔一个木糖就有一个乙酰基,由木聚糖乙酰转移酶(xylan acetyltransferase,XOAT)完成催化[31]。
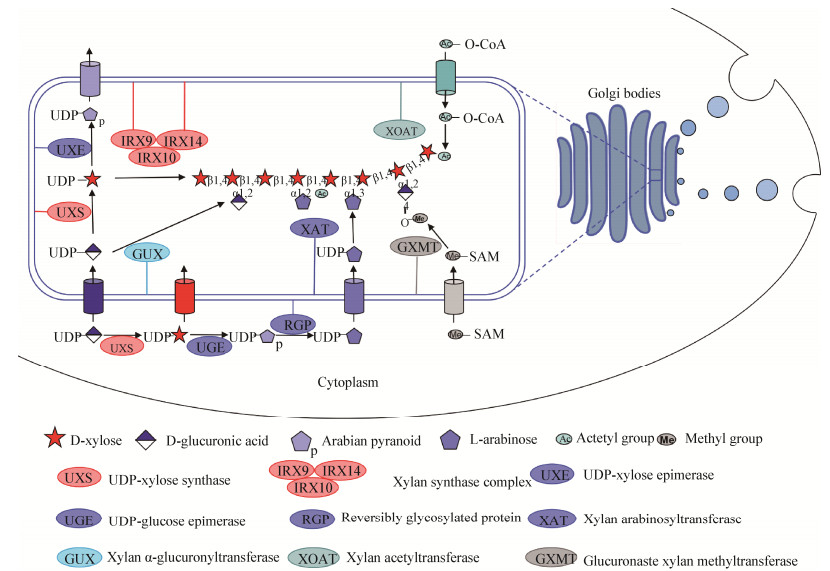
3 木聚糖降解酶系的多样性由于不同植物木聚糖组成与结构的复杂性,参与降解的微生物所产的木聚糖降解酶系也具有多样性(表 3),根据碳水化合物活性酶数据库(carbohydrate-active enzymes,CAZy)数据,来自古生菌、细菌和真菌的木聚糖降解酶系被归为不同的糖苷水解酶家族(GH family),通过水解机制可将木聚糖完全降解(图 3)。这些木聚糖降解酶系主要包括木聚糖主链降解酶类(内切木聚糖酶和β-木糖苷酶)和木聚糖侧链降解酶类。
| 木聚糖酶类型 Enzyme |
糖苷水解酶家族 GH family |
酶活类型 EC number |
底物特异性 Substrate specificity |
| 木聚糖主链降解酶 Xylan-backbone degradation enzymes |
|||
| 内切木聚糖酶 Endo-1, 4-β-xylanase |
GH5, 8, 10, 11, 30, 43, 51, 98 | EC 3.2.1.8 | GH5 is only active on GX/GAX; GH30 is only active on AX/GAX |
| 木糖苷酶 Exo-1, 4-β-xylosidase |
GH3, 39, 43, 52, 120 | EC 3.2.1.37 | Releases xylose from non-reducing ends |
| 木聚糖侧链降解酶 Xylan side-chain degradation enzymes |
|||
| 阿拉伯呋喃糖苷酶 α-L-arabinofuranosidase |
GH3, 43, 51, 54, 62 | EC3.2.1.55 | AXH-d is active on doubly substituted L-Araf residues; AXH-m is active on single substituted L-Araf residues |
| 葡萄糖醛酸酶 α-D-glucuronidase |
GH67, 115 | EC3.2.1.139 | GH67 is active on short xylooligosaccharides; GH115 is active on polymeric xylooligosaccharides |
| 乙酰木聚糖酯酶 Acetyl xylan esterase |
CE1-7, 12, 16 | EC3.1.1.72 | Liberates acetic acid from acetylated xylan; CE16 releases acetic acid from non-reducing ends |
| 阿魏酸酯酶 Ferulic acid esterase |
CE1, | EC3.1.1.73 | Liberates phenolic acids from xylan |
| 香豆酸酯酶 p-coumaric acid esterase |
CE15, | EC3.1.1.- | Liberates coumaric acid from xylan |
内切木聚糖酶(endo-1, 4-β-xylanase,EC 3.2.1.8)是木聚糖主链骨架降解的关键酶,能随机作用于木聚糖主链,将长链木聚糖水解成短链的低聚木糖,包括木二糖、木三糖、木四糖或更长的木寡糖。在CAZy数据库中木聚糖酶已归类为8个GH系列:GH5、GH8、GH10、GH11、GH30、GH43、GH51和GH98。据报道,GH5家族中第34亚家族的木聚糖酶类对阿拉伯糖基取代的木聚糖具有催化特异性[33]。GH8家族的木聚糖酶是唯一通过反转型催化机制进行催化的家族,其他家族为保留型催化机制。GH10家族的木聚糖酶对可溶性、具有侧链取代基的木聚糖存在较高的活性[34];而GH11家族的木聚糖酶对非取代的木聚糖催化活性较高[35]。另外,有报道证明GH10家族水解木聚糖底物产生的寡糖片段可以被GH11家族木聚糖酶进一步水解[36]。最新研究[37-39]发现,GH7和GH48家族的部分成员也具有木聚糖酶的水解功能,这与特定的催化活性架构密切相关,但具体的机制有待进一步的研究。
3.2 β-木糖苷酶β-木糖苷酶(exo-1, 4-β-xylosidase,EC 3.2.1.37)从木寡糖非还原端连续催化释放木糖残基,不能水解聚糖[40-41]。木糖苷酶主要存在于CAZy数据库的GH3、39、43、52、120家族中,具有保留型和反转型催化机制。许多木糖苷酶还存在转移酶活性,可以形成比底物分子量更高的产物,这表明相关酶类可应用于特定寡糖合成。
3.3 木聚糖侧链降解酶类由于木聚糖侧链取代基阻碍木聚糖主链降解酶的结合、识别与高效催化,因此需要在木聚糖侧链降解酶类的作用下除去取代基,从而促进木聚糖的高效降解。α-L-阿拉伯呋喃糖苷酶(α-L-arabinofuranosidases,EC3.2.1.55)催化水解木聚糖底物上的阿拉伯糖取代基,主要存在于GH3、43、51、54和62家族中[25]。催化木聚糖单取代基侧链的底物释放阿拉伯糖,相关酶类称作AXH-m,而从木聚糖双取代基侧链上催化释放阿拉伯糖基的酶类称为AXH-d[42]。α-D-葡萄糖醛酸酶(α-D-glucuronidases,EC3.2.1.139)催化木聚糖侧链4-O-甲基葡萄糖醛酸取代基的水解[33]。GH67家族葡萄糖醛酸酶更易于降解短链的葡萄糖醛酸取代的木寡糖[43],而GH115家族的酶对聚糖的活力更高[44]。另外,异质性木聚糖的有效降解还需要酯酶的参与,包括乙酰木聚糖酯酶(acetyl xylan esterases,EC3.1.1.72)水解木糖残基C-2或C-3位的O-乙酰侧链取代基、阿魏酸酯酶(ferulic acid esterases,EC3.1.1.73)和香豆酸酯酶(p-coumaric acid esterases,EC3.1.1.-)水解侧链阿拉伯糖C-2或C-5位和阿魏酰或对香豆酰残基之间的酯键。
3.4 木聚糖主链降解酶与侧链降解酶的协同作用迄今为止,木聚糖主链降解酶与侧链降解酶催化木聚糖的协同作用已被广泛研究。一方面由于不同糖苷水解酶家族的木聚糖酶存在不同的底物特异性,将不同家族木聚糖主链酶加入到反应体系中,促使催化反应加快并获得更高产量的目标产物[45-46]。另一方面,侧链降解酶将木聚糖侧链从主链骨架结构中去除,可促使木聚糖酶与底物主链骨架的可及性增加,反应速度加快。例如,GH115家族的葡萄糖醛酸酶去除了阻碍木聚糖酶结合的葡萄糖醛酸侧链,使产物木寡糖的产量大幅提高[47-48]。青霉GH11木聚糖酶XynC和黑曲霉的GH54阿拉伯呋喃糖苷酶AbfB在木寡糖的生产中可以高效协同降解不溶性小麦面粉阿拉伯木聚糖和黑麦面粉阿拉伯木聚糖[19]。乙酰取代基可能是内切木聚糖酶降解木聚糖的另一个障碍,被认为是植物对抗病原体体内木聚糖降解酶的防御机制[49]。对于热预处理和二甲基亚砜(dimethyl sulfoxide,DMSO)萃取产生的乙酰化的木聚糖原料,将木聚糖酶与乙酰木聚糖酯酶一起加入木聚糖降解反应体系中,两种酶能够表现出明显的协同作用[50]。另外,随着基因组学技术的快速发展,人们还发现了作用于木聚糖的裂解多糖单加氧酶家族(AA14),其与GH11家族木聚糖酶一起可高效降解木聚糖[51]。
4 木聚糖高效降解酶系的设计与定制目前,木聚糖高效降解酶系的相关研究主要聚焦天然生境中微生物群落分泌的降解酶系、优势微生物的降解酶系及针对特定的目标底物定制绿色高效降解酶系。
4.1 微生物群落产生的混合酶系木质纤维素资源可被天然生境中复杂的微生物群落高效降解转化,这些微生物共同分泌高效降解酶类参与降解转化。本实验室基于整合宏组学技术来认识天然腐生生境微生物区系及其功能基因,发现在玉米秸秆堆肥微生物群落里的主导菌属是真菌的嗜热丝孢菌属、细菌的嗜热多孢菌属和喜热裂孢菌属[52]。这3种菌属都具有耐热性和木质纤维素降解能力,其中嗜热丝孢菌属和嗜热多孢菌属能够分泌木聚糖酶进行异质木聚糖的高效降解,喜热裂孢菌属是细菌中主要的纤维素降解者,也分泌高效的嗜热木聚糖主链降解酶和侧链降解酶参与半纤维素类的降解转化[52]。此外,从天然生境中提取的酶系可与外源酶系协同作用。例如,从牛瘤胃中获得的内源性酶与添加到饲料中的外源性真菌酶相互协同作用,导致可溶性纤维素、木聚糖和玉米青贮物的水解显著增加[53]。近年来,人们根据代谢功能将分离出的微生物群落进行系统组合,其产生混合酶系的降解潜力几乎可以媲美完整菌群的降解能力[54]。
4.2 优势微生物的降解酶系由于不同种类植物进化途径及生长环境迥异,使得植物细胞壁多糖的组成及结构存在明显异质;而微生物与植物共进化形成了多种多样的降解利用策略[55-56]。Aspergillus niger An76全基因组测序分析发现多种木聚糖降解同工酶,有5个木聚糖内切酶基因、2个β-木糖苷酶基因、9个阿拉伯呋喃糖苷酶基因、1个葡萄糖醛酸酶基因、9个阿魏酸酯酶基因以及2个乙酰木聚糖酯酶基因,具有彻底降解复杂木聚糖的能力[57]。本实验室利用功能蛋白质组学方法系统分析了胞外及胞内蛋白质组的动态变化,发现木聚糖主链降解酶系要先于侧链降解酶系的表达[58],通过降解产物动态寡糖谱的展示,发现不同家族木聚糖酶(GH10和GH11)产物谱的特异性都存在明显差异[35]。不同家族木聚糖降解同工酶对底物结合模式、产物寡糖谱以及表达调控中对不同底物的响应机制存在明显差异,这些因素都会影响木聚糖的高效转化。
4.3 基于特定底物设计绿色高效的降解酶系随着不同植物木聚糖精细结构的深入研究及酶分子结构功能关系的阐明,针对特定底物种类(如大麦、小麦、燕麦及玉米等)设计并定制高效降解酶系成为可能。研究表明特制酶系统一般可由核心酶和辅助酶组成,使用系统实验设计来优化其化学计量比。Meyer等针对不同理化性质或不同预处理的农业残留物,通过酶活性的鉴定(主要和次要酶)设计并优化了生物质加工所需酶活性最小数量的组合[59]。Kim等通过优化酶的浓度提高降解速率,针对酸预处理的稻草(56.9%葡聚糖,0.0%木聚糖,24.8%木质素)和碱预处理的稻草(47.6%葡聚糖,13.1%木聚糖,15.1%木质素)定制酶系,高效降解酸处理稻草的酶系比例内切纤维素酶E3:低浓度的外切酶Cel5H:木聚糖酶Xyn10C是23.031:6.969:0 (mg/g,质量比);而高效降解碱处理稻草的酶系比例E3:Cel5H:Xyn10C是7.962:9.012:13.029 (mg/g,质量比)[60]。基于优势微生物分泌酶系分析降解不同底物时的限速步骤,有目标地定量添加经过工程化改造过的特定酶类,可以构建绿色低成本高效木聚糖降解复合酶系。另外,Huang等研究了芽孢杆菌NG80-2中的木聚糖酶、阿拉伯呋喃糖苷酶和木糖苷酶3种酶之间的协同作用,3种酶的协同作用可以降解木聚糖底物使单糖产量提升[61]。
酶系定制添加的酶组分常常与原有酶系的性质不一致,因此常利用蛋白质工程来改善酶的生化性质。伴随着生物大数据时代的来临,蛋白质工程技术的不断创新,杂合酶的设计迎来了发展的新趋势[62]。Mckee等分析了GH43家族阿拉伯呋喃糖苷酶HiAXHd3活性中心氨基酸与底物的结构,在酶的活性中心远端亚位点引入Y166A的突变,使得木聚糖酶活力显著增加,并保留了原有的阿拉伯呋喃糖苷酶活性[63]。很多文献表明通过设计酶分子的N末端区域可以提高木聚糖酶稳定性[64-65]。通过B因子分析和多序列比对的方法指导进行突变,获得的4种突变体的热稳定性都有所增强[66]。Wu等通过对GH11家族的木聚糖酶远端亚位点2个氨基酸的替换,使得酶分子的活性和温度稳定性都有所提高[67]。另外,新计算方法的引入也为酶的功能设计带来了巨大的希望[68-69]。Wang等考虑到多种酶的生产成本,通过将木聚糖酶与阿魏酸酯酶融合,构建了嵌合的双功能木聚糖酶/阿魏酸酯酶rXyn10A/Fae1A,阿魏酸和木寡糖的产量都有所提高[70]。另外,McClendon等以纤维小体的结构为模板,开发了4种不同的木聚糖小体,每种至少包含3种不同的半纤维素酶活性,并且在降解小麦阿拉伯木聚糖时,这些木聚糖小体在还原糖和阿魏酸的释放方面都优于游离酶系统[71]。将木聚糖降解酶系统和酵母表面展示模块相结合,构建酵母表面的木聚糖小体,从而增强半纤维素底物在酵母中的糖化作用[72]。虽然构建木聚糖小体已经有很多尝试,但是更多的研究需要进行,依据木聚糖的结构类型构建特定的木聚糖酶复合体,从而实现木聚糖的高效降解,是目前完成木聚糖高效降解的新挑战。
5 木聚糖降解酶系的精细定制与应用前景 5.1 能源行业木质纤维素原料降解转化不仅可促进自然界碳素循环,而且可增加能源供给并减轻环境污染,因而成为国内外学者的研究热点[73]。复杂的木聚糖是木质纤维素原料的外层抗降解屏障,其高效彻底降解至少需要20个家族7种糖苷水解酶及酯酶共同作用才能完成[32]。自然界已进化出高效降解转化木质纤维素的相关菌株,例如Beri等分离了一株能够高效降解GAX的细菌,通过高温菌共培养及其定制纯的侧链酶,可使玉米纤维转化为生物乙醇的产率由53%±3%提高到76%±1%[74]。然而仍有部分GAX没有被利用,因此,深入研究GAX残余部分的精细结构,定制高效降解相关木聚糖的酶(系)仍是下一步研究的重点。此外,胞内C5与C6的单糖同步快速利用也是生物能源行业的难点和挑战,这需要利用合成生物学新技术完成工程菌株的构建或共培养来提高同步乙醇的产率与效率。
5.2 造纸工业木聚糖酶从20世纪80年代开始就广泛应用于造纸行业中,主要应用在生物漂白等相关技术,利用木聚糖酶降解纤维素表面的半纤维素,从而增加纤维纸张的白度。生物漂白应用的环境是碱性环境,而多数天然糖苷水解酶的最适pH一般是酸中性,因此,有目的地定制与改造相关木聚糖适应碱性环境是造纸用酶筛选与改造的方向。Wu等通过对木聚糖酶表面极性氨基酸有目的的设计,在耐热木聚糖酶TlXynA的基础上使其具有耐碱性的能力,因而引起国内同行的关注[75]。
5.3 食品行业木寡糖是木聚糖降解的中间产物,作为新兴益生元可以促进对人类和动物肠道益生菌的快速增长,因而具有明显的益生作用[76-79]。然而,现有方法多利用粗酶系来生产木寡糖,获得的成分也多为混合物,并且不易控制产品质量等,因此,基于特定底物结构分析精确定制绿色高效相关酶类来提高降解效率并降低生产成本,这仍是该领域的研究挑战,需要引入最新基因编辑手段对特定微生物胞外酶系统进行系统改造,才能完成特定高效酶系的定制。
6 展望木聚糖高效降解研究已有较长的历史,由于其结构复杂性仍然需要深入研究,特别对我国这一粮食利用大国,大宗粮食加工的副产物主要是半纤维素的残余,比如啤酒发酵后的麦糟残余、燕麦及糙米加工副产物等,这些物质一般作为饲料,未进一步深入加工利用。然而,这些物质降解转化为木寡糖、木糖、阿拉伯糖等新兴益生元,可以应用于功能食品、医疗和制药行业。由于木聚糖底物的降解效率与定制寡糖产物的产量及纯度都达不到预期,形成了绿色高效转化的瓶颈,因此,对特定的底物结构进行精准分析、基于产品的需求定制微生物高效降解酶组分及其组合、构建低成本的相应酶系生产微生物工程菌株仍应是重要的研究方向。而且随着组学技术的快速发展,生物大数据时代提高了理性设计的速度,利用高通量基因组技术从微生物资源中筛选全新木聚糖酶系,进而优化定制绿色高效的木聚糖降解酶系成为可能,从而可以推动相关产业的快速发展。
| [1] |
Konishi T, Nakata I, Miyagi Y, et al. Extraction of β-1, 3 xylan from green seaweed, Caulerpa lentillifera[J]. Journal of Applied Glycoscience, 2012, 59(4): 161-163. DOI:10.5458/jag.jag.JAG-2011_025 |
| [2] |
Viana AG, Noseda MD, Gonçalves AG, et al. β-D-(1→4), β-D-(1→3) 'mixed linkage' xylans from red seaweeds of the order Nemaliales and Palmariales[J]. Carbohydrate Research, 2011, 346(8): 1023-1028. DOI:10.1016/j.carres.2011.03.013 |
| [3] |
Scheller HV, Ulvskov P. Hemicelluloses[J]. Annual Review of Plant Biology, 2010, 61(1): 263-289. DOI:10.1146/annurev-arplant-042809-112315 |
| [4] |
Peng YY, Wu SB. The structural and thermal characteristics of wheat straw hemicellulose[J]. Journal of Analytical and Applied Pyrolysis, 2010, 88(2): 134-139. DOI:10.1016/j.jaap.2010.03.006 |
| [5] |
Zhou XW, Li WJ, Mabon R, et al. A critical review on hemicellulose pyrolysis[J]. Energy Technology, 2017, 5(1): 52-79. |
| [6] |
Cordeiro LM, de Almeida CP, Iacomini M. Unusual linear polysaccharides: (1→5)-α-L-Arabinan, (1→3)-(1→4)-α-D- glucan and (1→4)-β-D-xylan from pulp of buriti (Mauritia flexuosa), an edible palm fruit from the Amazon region[J]. Food Chemistry, 2015, 173: 141-146. DOI:10.1016/j.foodchem.2014.10.020 |
| [7] |
Ebringerová A, Hromádková Z, Heinze T. Hemicellulose[A]//Heinze T. Polysaccharides I[M]. Berlin: Springer, 2005: 1-67
|
| [8] |
Rogowski A, Briggs JA, Mortimer JC, et al. Glycan complexity dictates microbial resource allocation in the large intestine[J]. Nature Communications, 2015, 6: 7481. DOI:10.1038/ncomms8481 |
| [9] |
Deutschmann R, Dekker RFH. From plant biomass to bio-based chemicals: latest developments in xylan research[J]. Biotechnology Advances, 2012, 30(6): 1627-1640. |
| [10] |
Busse-Wicher M, Gomes TCF, Tryfona T, et al. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana[J]. The Plant Journal, 2014, 79(3): 492-506. |
| [11] |
Chong SL, Virkki L, Maaheimo H, et al. O-Acetylation of glucuronoxylan in Arabidopsis thaliana wild type and its change in xylan biosynthesis mutants[J]. Glycobiology, 2014, 24(6): 494-506. DOI:10.1093/glycob/cwu017 |
| [12] |
Li XF, Jackson P, Rubtsov DV, et al. Development and application of a high throughput carbohydrate profiling technique for analyzing plant cell wall polysaccharides and carbohydrate active enzymes[J]. Biotechnology for Biofuels, 2013, 6(1): 94. |
| [13] |
Coughlan MP, Tuohy MG, Filho EXF, et al. Enzymological aspects of microbial hemicellulases with emphasis on fungal systems[A]//Coughlan MP, Hazlewood GP. Hemicellulose and Hemicellulases[M]. London: Portland Press, 1993: 53-84
|
| [14] |
Stephen AM. Other plant polysaccharides[A]//Aspinall GO. The Polysaccharides[M]. New York: Academic Press, 1983: 97-103
|
| [15] |
Andersson R, Åman P. Cereal arabinoxylan: occurrence, structure and properties[A]//McCleary BV, Prosky L. Advanced Dietary Fibre Technology[M]. Oxford: Blackwell Science, 2000: 299-314
|
| [16] |
De O Buanafina MM. Feruloylation in grasses: current and future perspectives[J]. Molecular Plant, 2009, 2(5): 861-872. DOI:10.1093/mp/ssp067 |
| [17] |
Rennie EA, Scheller HV. Xylan biosynthesis[J]. Current Opinion in Biotechnology, 2014, 26: 100-107. |
| [18] |
Liu L, Gong WL, Sun XM, et al. Extracellular enzyme composition and functional characteristics of Aspergillus niger AN-76 induced by food processing byproducts and based on integrated functional omics[J]. Journal of Agricultural and Food Chemistry, 2018, 66(5): 1285-1295. DOI:10.1021/acs.jafc.7b05164 |
| [19] |
Gonçalves TA, Damásio ARL, Segato F, et al. Functional characterization and synergic action of fungal xylanase and arabinofuranosidase for production of xylooligosaccharides[J]. Bioresource Technology, 2012, 119: 293-299. DOI:10.1016/j.biortech.2012.05.062 |
| [20] |
Fangel JU, Ulvskov P, Knox JP, et al. Cell wall evolution and diversity[J]. Frontiers in Plant Science, 2012, 3: 152. |
| [21] |
Smith PJ, Wang HT, York WS, et al. Designer biomass for next-generation biorefineries: leveraging recent insights into xylan structure and biosynthesis[J]. Biotechnology for Biofuels, 2017, 10(1): 286. |
| [22] |
Lee C, O'Neill MA, Tsumuraya Y, et al. The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity[J]. Plant and Cell Physiology, 2007, 48(11): 1624-1634. |
| [23] |
Brown DM, Goubet F, Wong VW, et al. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis[J]. The Plant Journal, 2007, 52(6): 1154-1168. DOI:10.1111/j.1365-313X.2007.03307.x |
| [24] |
Peña MJ, Zhong RQ, Zhou GK, et al. Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis[J]. The Plant Cell, 2007, 19(2): 549-563. |
| [25] |
Lombard V, Ramulu HG, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013[J]. Nucleic Acids Research, 2014, 42(D1): D490-495. DOI:10.1093/nar/gkt1178 |
| [26] |
Rennie EA, Hansen SF, Baidoo EEK, et al. Three members of the Arabidopsis glycosyltransferase family 8 are xylan glucuronosyltransferases[J]. Plant Physiology, 2012, 159(4): 1408-1417. |
| [27] |
Mortimer JC, Miles GP, Brown DM, et al. Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(40): 17409-17414. DOI:10.1073/pnas.1005456107 |
| [28] |
Bromley JR, Busse-Wicher M, Tryfona T, et al. GUX 1 and GUX 2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns[J]. The Plant Journal, 2013, 74(3): 423-434. DOI:10.1111/tpj.12135 |
| [29] |
Zeng W, Jiang N, Nadella R, et al. A glucurono (arabino) xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively[J]. Plant Physiology, 2010, 154(1): 78-97. |
| [30] |
Rautengarten C, Ebert B, Herter T, et al. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis[J]. The Plant Cell, 2011, 23(4): 1373-1390. |
| [31] |
Xiong GY, Dama M, Pauly M. Glucuronic acid moieties on xylan are functionally equivalent to O-acetyl-substituents[J]. Molecular Plant, 2015, 8(7): 1119-1121. DOI:10.1016/j.molp.2015.02.013 |
| [32] |
Gong WL, Wang LS, Zhang HQ, et al. Diverse synthetases and fungi degradation enzymes for the polysaccharides of plant cell walls[J]. Biotechnology Bulletin, 2015, 31(4): 149-165. (in Chinese) 公维丽, 王禄山, 张怀强, 等. 植物细胞壁多糖合成酶系及真菌降解酶系[J]. 生物技术通报, 2015, 31(4): 149-165. |
| [33] |
Karlsson EN, Schmitz E, Linares-Pastén JA, et al. Endo-xylanases as tools for production of substituted xylooligosaccharides with prebiotic properties[J]. Applied Microbiology and Biotechnology, 2018, 102(21): 9081-9088. DOI:10.1007/s00253-018-9343-4 |
| [34] |
Biely P, Singh S, Puchart V. Towards enzymatic breakdown of complex plant xylan structures: state of the art[J]. Biotechnology Advances, 2016, 34(7): 1260-1274. DOI:10.1016/j.biotechadv.2016.09.001 |
| [35] |
Gong WL, Zhang HQ, Tian L, et al. Determination of the modes of action and synergies of xylanases by analysis of xylooligosaccharide profiles over time using fluorescence-assisted carbohydrate electrophoresis[J]. Electrophoresis, 2016, 37(12): 1640-1650. DOI:10.1002/elps.201600041 |
| [36] |
Kolenová K, Vršanská M, Biely P. Purification and characterization of two minor endo-β-1, 4-xylanases of Schizophyllum commune[J]. Enzyme and Microbial Technology, 2005, 36(7): 903-910. DOI:10.1016/j.enzmictec.2005.01.006 |
| [37] |
Chu YD, Hao ZZ, Wang KK, et al. The GH10 and GH48 dual-functional catalytic domains from a multimodular glycoside hydrolase synergize in hydrolyzing both cellulose and xylan[J]. Biotechnology for Biofuels, 2019, 12: 279. DOI:10.1186/s13068-019-1617-2 |
| [38] |
Han C, Yang RR, Sun YX, et al. Identification and characterization of a novel hyperthermostable bifunctional cellobiohydrolase-xylanase enzyme for synergistic effect with commercial cellulase on pretreated wheat straw degradation[J]. Frontiers in Bioengineering and Biotechnology, 2020, 8: 296. DOI:10.3389/fbioe.2020.00296 |
| [39] |
Hua CY, Li WG, Han W, et al. Characterization of a novel thermostable GH7 endoglucanase from Chaetomium thermophilum capable of xylan hydrolysis[J]. International Journal of Biological Macromolecules, 2018, 117: 342-349. DOI:10.1016/j.ijbiomac.2018.05.189 |
| [40] |
Yan QJ, Wang L, Jiang ZQ, et al. A xylose-tolerant β-xylosidase from Paecilomyces thermophila: characterization and its co-action with the endogenous xylanase[J]. Bioresource Technology, 2008, 99(13): 5402-5410. DOI:10.1016/j.biortech.2007.11.033 |
| [41] |
Knob A, Terrasan CRF, Carmona EC. β-Xylosidases from filamentous fungi: an overview[J]. World Journal of Microbiology and Biotechnology, 2010, 26(3): 389-407. |
| [42] |
Lagaert S, Pollet A, Courtin CM, et al. β-xylosidases and α-L-arabinofuranosidases: accessory enzymes for arabinoxylan degradation[J]. Biotechnology Advances, 2014, 32(2): 316-332. |
| [43] |
Golan G, Shallom D, Teplitsky A, et al. Crystal structures of Geobacillus stearothermophilus α-glucuronidase complexed with its substrate and products mechanistic implications[J]. Journal of Biological Chemistry, 2004, 279(4): 3014-3024. |
| [44] |
McKee LS, Sunner H, Anasontzis GE, et al. A GH115 α-glucuronidase from Schizophyllum commune contributes to the synergistic enzymatic deconstruction of softwood glucuronoarabinoxylan[J]. Biotechnology for Biofuels, 2016, 9(1): 2. |
| [45] |
Gonçalves GAL, Takasugi Y, Jia LL, et al. Synergistic effect and application of xylanases as accessory enzymes to enhance the hydrolysis of pretreated bagasse[J]. Enzyme and Microbial Technology, 2015, 72: 16-24. DOI:10.1016/j.enzmictec.2015.01.007 |
| [46] |
Malgas S, Pletschke BI. The effect of an oligosaccharide reducing-end xylanase, BhRex8A, on the synergistic degradation of xylan backbones by an optimised xylanolytic enzyme cocktail[J]. Enzyme and Microbial Technology, 2019, 122: 74-81. DOI:10.1016/j.enzmictec.2018.12.010 |
| [47] |
Rosa L, Ravanal MC, Mardones W, et al. Characterization of a recombinant α-glucuronidase from Aspergillus fumigatus[J]. Fungal Biology, 2013, 117(5): 380-387. DOI:10.1016/j.funbio.2013.04.002 |
| [48] |
Wang LL, Shi H, Xu BY, et al. Characterization of Thermotoga thermarum DSM 5069 α-glucuronidase and synergistic degradation of xylan[J]. BioResources, 2016, 11(3): 5767-5779. |
| [49] |
Tenkanen M, Vršanská M, Siika-aho M, et al. Xylanase XYN IV from Trichoderma reesei showing exo- and endo-xylanase activity[J]. FEBS Journal, 2013, 280(1): 285-301. DOI:10.1111/febs.12069 |
| [50] |
Huang YC, Chen GH, Chen YF, et al. Heterologous expression of thermostable acetylxylan esterase gene from Thermobifida fusca and its synergistic action with xylanase for the production of xylooligosaccharides[J]. Biochemical and Biophysical Research Communications, 2010, 400(4): 718-723. DOI:10.1016/j.bbrc.2010.08.136 |
| [51] |
Couturier M, Ladevèze S, Sulzenbacher G, et al. Lytic xylan oxidases from wood-decay fungi unlock biomass degradation[J]. Nature Chemical Biology, 2018, 14(3): 306-310. DOI:10.1038/nchembio.2558 |
| [52] |
Zhang LL, Ma HX, Zhang HQ, et al. Thermomyces lanuginosus is the dominant fungus in maize straw composts[J]. Bioresource Technology, 2015, 197: 266-275. DOI:10.1016/j.biortech.2015.08.089 |
| [53] |
Morgavi DP, Beauchemin KA, Nsereko VL, et al. Synergy between ruminal fibrolytic enzymes and enzymes from Trichoderma longibrachiatum[J]. Journal of Dairy Science, 2000, 83(6): 1310-1321. DOI:10.3168/jds.S0022-0302(00)74997-6 |
| [54] |
Puentes-Téllez PE, Salles JF. Construction of effective minimal active microbial consortia for lignocellulose degradation[J]. Microbial Ecology, 2018, 76(2): 419-429. DOI:10.1007/s00248-017-1141-5 |
| [55] |
Gong WL, Zhang HQ, Liu SJ, et al. Comparative secretome analysis of Aspergillus niger, Trichoderma reesei, and Penicillium oxalicum during solid-state fermentation[J]. Applied Biochemistry and Biotechnology, 2015, 177(6): 1252-1271. |
| [56] |
Champreda V, Mhuantong W, Lekakarn H, et al. Designing cellulolytic enzyme systems for biorefinery: from nature to application[J]. Journal of Bioscience and Bioengineering, 2019, 128(6): 637-654. DOI:10.1016/j.jbiosc.2019.05.007 |
| [57] |
Gong WL, Cheng Z, Zhang HQ, et al. Draft genome sequence of Aspergillus niger strain An76[J]. Genome Announcements, 2016, 4(1): e01700-15. |
| [58] |
Gong WL, Dai L, Zhang HQ, et al. A highly efficient xylan-utilization system in Aspergillus niger AN76: a functional-proteomics study[J]. Frontiers in Microbiology, 2018, 9: 430. DOI:10.3389/fmicb.2018.00430 |
| [59] |
Meyer AS, Rosgaard L, Sørensen HR. The minimal enzyme cocktail concept for biomass processing[J]. Journal of Cereal Science, 2009, 50(3): 337-344. DOI:10.1016/j.jcs.2009.01.010 |
| [60] |
Kim IJ, Jung JY, Lee HJ, et al. Customized optimization of cellulase mixtures for differently pretreated rice straw[J]. Bioprocess and Biosystems Engineering, 2015, 38(5): 929-937. |
| [61] |
Huang D, Liu J, Qi YF, et al. Synergistic hydrolysis of xylan using novel xylanases, β-xylosidases, and an α-L-arabinofuranosidase from Geobacillus thermodenitrificans NG80-2[J]. Applied Microbiology and Biotechnology, 2017, 101(15): 6023-6037. DOI:10.1007/s00253-017-8341-2 |
| [62] |
Zhang Q, Wu XY, Jiang XK, et al. Trend of hybrid enzyme design in the big data era[J]. Chinese Journal of Biotechnology, 2018, 34(7): 1033-1045. (in Chinese) 张群, 吴秀芸, 蒋绪恺, 等. 大数据时代杂合酶的设计及其新趋势[J]. 生物工程学报, 2018, 34(7): 1033-1045. |
| [63] |
McKee LS, Peña MJ, Rogowski A, et al. Introducing endo-xylanase activity into an exo-acting arabinofuranosidase that targets side chains[J]. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(17): 6537-6542. DOI:10.1073/pnas.1117686109 |
| [64] |
Wang YW, Fu Z, Huang HQ, et al. Improved thermal performance of Thermomyces lanuginosus GH11 xylanase by engineering of an N-terminal disulfide bridge[J]. Bioresource Technology, 2012, 112: 275-279. DOI:10.1016/j.biortech.2012.02.092 |
| [65] |
Cheng YS, Chen CC, Huang CH, et al. Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum[J]. Journal of Biological Chemistry, 2014, 289(16): 11020-11028. DOI:10.1074/jbc.M114.550905 |
| [66] |
Han NY, Ma Y, Mu YL, et al. Enhancing thermal tolerance of a fungal GH11 xylanase guided by B-factor analysis and multiple sequence alignment[J]. Enzyme and Microbial Technology, 2019, 131: 109422. DOI:10.1016/j.enzmictec.2019.109422 |
| [67] |
Wu XY, Tian ZN, Jiang XK, et al. Enhancement in catalytic activity of Aspergillus niger XynB by selective site-directed mutagenesis of active site amino acids[J]. Applied Microbiology and Biotechnology, 2018, 102(1): 249-260. |
| [68] |
Goldenzweig A, Goldsmith M, Hill SE, et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability[J]. Molecular Cell, 2016, 63(2): 337-346. |
| [69] |
Khersonsky O, Lipsh R, Avizemer Z, et al. Automated design of efficient and functionally diverse enzyme repertoires[J]. Molecular Cell, 2018, 72(1): 178-186. |
| [70] |
Wang RN, Yang JS, Jang JM, et al. Efficient ferulic acid and xylo-oligosaccharides production by a novel multi-modular bifunctional xylanase/feruloyl esterase using agricultural residues as substrates[J]. Bioresource Technology, 2020, 297: 122487. DOI:10.1016/j.biortech.2019.122487 |
| [71] |
McClendon SD, Mao ZC, Shin HD, et al. Designer xylanosomes: protein nanostructures for enhanced xylan hydrolysis[J]. Applied Biochemistry and Biotechnology, 2012, 167(3): 395-411. |
| [72] |
Srikrishnan S, Chen W, da Silva NA. Functional assembly and characterization of a modular xylanosome for hemicellulose hydrolysis in yeast[J]. Biotechnology and Bioengineering, 2013, 110(1): 275-285. DOI:10.1002/bit.24609 |
| [73] |
Lynd LR, Liang XY, Biddy MJ, et al. Cellulosic ethanol: status and innovation[J]. Current Opinion in Biotechnology, 2017, 45: 202-211. DOI:10.1016/j.copbio.2017.03.008 |
| [74] |
Beri D, York WS, Lynd LR, et al. Development of a thermophilic coculture for corn fiber conversion to ethanol[J]. Nature Communications, 2020, 11(1): 1937. DOI:10.1038/s41467-020-15704-z |
| [75] |
Wu XY, Zhang Q, Zhang LZ, et al. Insights into the role of exposed surface charged residues in the alkali-tolerance of GH11 xylanase[J]. Frontiers in Microbiology, 2020, 11: 872. DOI:10.3389/fmicb.2020.00872 |
| [76] |
Aachary AA, Gobinath D, Srinivasan K, et al. Protective effect of xylooligosaccharides from corncob on 1, 2-dimethylhydrazine induced colon cancer in rats[J]. Bioactive Carbohydrates and Dietary Fibre, 2015, 5(2): 146-152. DOI:10.1016/j.bcdf.2015.03.004 |
| [77] |
Krishna G, Divyashri G, Prapulla SG, et al. A combination supplement of fructo- and xylo-oligosaccharides significantly abrogates oxidative impairments and neurotoxicity in maternal/fetal milieu following gestational exposure to acrylamide in rat[J]. Neurochemical Research, 2015, 40(9): 1904-1918. DOI:10.1007/s11064-015-1687-x |
| [78] |
Yang JP, Summanen PH, Henning SM, et al. Xylooligosaccharide supplementation alters gut bacteria in both healthy and prediabetic adults: a pilot study[J]. Frontiers in Physiology, 2015, 6: 216. |
| [79] |
Lin SH, Chou LM, Chien YW, et al. Prebiotic effects of xylooligosaccharides on the improvement of microbiota balance in human subjects[J]. Gastroenterology Research and Practice, 2016, 2016: 5789232. |
 2020, Vol. 47
2020, Vol. 47