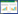扩展功能
文章信息
- 石小玉, 张宁, 宁娜, 员超, 倪金凤
- SHI Xiao-Yu, ZHANG Ning, NING Na, YUN Chao, NI Jin-Feng
- 一株培菌白蚁后肠木聚糖降解细菌的分离与鉴定
- Isolation and identification of xylan-degradation bacteria from hindgut of fungus-growing termite
- 微生物学通报, 2016, 43(3): 504-509
- Microbiology China, 2016, 43(3): 504-509
- 10.13344/j.microbiol.china.150457
-
文章历史
- 收稿日期: 2015-06-12
- 接受日期: 2015-08-18
- 优先数字出版日期(www.cnki.net): 2015-09-17
木质纤维素中的半纤维素含量约占20%-30%,是自然界中含量仅次于纤维素的成分。半纤维素中含量最丰富的成分是木聚糖[1],木聚糖是植物细胞中一种主要的结构多糖,是自然界含量位居第二位的多糖,约占可再生有机碳源的1/3[2]。木聚糖酶是将木聚糖降解成低聚木糖和木糖的一类水解酶。已知产木聚糖酶的生物很多,如细菌、酵母、放线菌、真菌等微生物[3, 4, 5, 6],另外蜗牛、海洋藻类、陆生植物组织、反刍动物瘤胃及白蚁也具有木聚糖酶[1, 4, 7]。白蚁是热带和亚热带地区木质纤维素的重要分解者[8]。白蚁约有2 800多种,不同的白蚁种类,其木质纤维素酶分布及降解机制不同[4]。截止目前研究,一般认为白蚁自身不产木聚糖酶。在低等白蚁后肠存在由其共生原生动物分泌的木聚糖酶[9],最近报道从低等白蚁中分离到具有木聚糖等多糖降解活性的细菌[10]。高等白蚁肠道不含原生动物,其共生的细菌可能在木聚糖降解中发挥重要作用。宏基因组研究分析发现高等食木白蚁后肠有丰富的木质纤维素降解基因[11],通过构建后肠微生物fosmid文库从高等白蚁后肠中获得具有木聚糖基因的克隆[12, 13]。黄翅大白蚁是广泛分布于我国南方的高等培菌白蚁,本文以黄翅大白蚁后肠为材料,通过常规的平板活性筛选方法,分离到一株具有木聚糖降解活性的微生物,进一步对其进行了鉴定和初步的酶活性分析。
1 材料与方法 1.1 材料 1.1.1 样品:黄翅大白蚁取自湖南省耒阳市木兰村,采样后立即冻存于-80 ℃,所用感受态细胞为自制大肠杆菌感受态Escherichia coli DH5α;测序载体为质粒pMD18T vector (Takara公司)。 1.1.2 培养基及主要试剂:基本盐培养基[14]:磷酸二氢钾0.20 g/L,氯化铵0.25 g/L,氯化钾0.50 g/L,二水氯化钙0.15 g/L,氯化钠1.00 g/L,醋酸钠2.46 g/L,六水氯化镁0.62 g/L,硫酸钠2.84 g/L,4-羟乙基哌嗪乙磺酸(HEPES) (pH 6.8) 10 mmol/L,微量元素溶液1 mL,维生素溶液1 mL。其中,微量元素溶液所含成分为:四水氯化亚铁1.50 g/L,六水氯化钴190 mg/L,四水氯化锰100 mg/L,氯化锌70 mg/L,锰酸6 mg/L,二水钼酸钠 36 mg/L,一水氯化镍24 mg/L,一水氯化钙2 mg/L,盐酸(25%,体积比) 10 mL;维生素溶液中的成分为(mg/L):4-氨基苯甲酸40,生物素10,烟酸100,泛酸钙50;微量元素溶液和维生素溶液过滤除菌后于4 ℃保存。选择培养基是在去除醋酸钠的基本盐培养基中加入0.2%的桦木木聚糖。产酶培养基是在选择培养基基础上加入0.1%蛋白胨。3,5-二硝基水扬酸(DNS)试剂配制见参考文献[15]。Taq酶购自Trans公司;桦木木聚糖和燕麦木聚糖购自Sigma公司;蛋白胨购自Oxoid公司;引物合成及PCR产物测序由北京六合华大基因科技股份有限公司完成。 1.2 样品处理及菌株的分离 1.2.1 样品处理:在无菌条件下,将50只黄翅大白蚁工蚁在超净台解剖,将获得的后肠放置于1.5 mL离心管,并用灭菌的研磨棒研磨至无可见颗粒,作为后肠微生物培养样品。以0.5%接种量接种黄翅大白蚁后肠微生物样品至5 mL选择培养基中,30 ℃、150 r/min培养2 d至木聚糖降解较完全后,以2%接种量转接入5 mL新的选择培养基中,于30 ℃、150 r/min进行第2次富集培养。 1.2.2 菌落筛选:第2次富集的样品用选择培养基进行梯度稀释,取稀释104和105倍的培养液100 μL分别涂布于选择培养基平板上,30 ℃培养3 d,再挑取单菌落至新的选择培养基上,30 ℃继续培养,3 d后通过平板上可见的透明区域并结合刚果红活性染色挑选克隆。刚果红染色具体方法为:70%乙醇洗去菌落,用0.3%的刚果红溶液染色20 min,然后用1 mol/L NaCl脱色20 min。挑选水解圈大的菌落进行划线分离,并且进行选择平板筛选和刚果红活性验证,观察水解圈的大小,选择木聚糖活性高的菌株。 1.3 菌株鉴定 1.3.1 革兰氏染色与镜检:革兰氏染色方法见参考文献[16]。 1.3.2 基因组DNA提取和16S rRNA基因扩增及序列分析:用十六烷基三甲基溴化铵(CTAB)法提取基因组DNA。具体步骤如下:在菌体中加入200 μL的裂解缓冲液(50 mmol/L Tris-HCl,pH 8.0;25 mmol/L EDTA;3% SDS;1% CTAB,1.2% PVP),65 ℃水浴50 min;然后加入800 μL CTAB提取缓冲液(10 mmol/L Tris-HCl,pH 8.0;1 mmol/L EDTA;0.3 mmol/L醋酸钠;2% CTAB,1.2% PVP)。每5 min轻轻振荡20 s,重复4次后,8 000 r/min离心20 min取上清;加入等体积的酚:氯仿溶液,混匀后8 000 r/min离心10 min取上清,重复该步骤1-2次至不出现蛋白层为止;之后取上清;-20 ℃沉淀大于1 h后离心弃上清,用70%乙醇洗涤沉淀2次后,置于室温中干燥15 min,加入30-50 μL ddH2O,作为提取的该菌株的基因组。16S rRNA基因扩增引物分别是27f (5′-AGAGTTTGATCCTGGCTCAG-3′)和1492R (5′-GGTTACCTTGTTACGACTT-3′)。PCR反应体系:0.2 mL EP管中加入1 μL DNA,10 μmol/L上下游引物各1 μL,10×PCR buffer 5 μL,2.5 mmol/L dNTPs 4 μL,5 U/μL Taq酶0.5 μL,加ddH2O使终体积为50 μL,混合均匀。反应条件:94 ℃ 5 min;94 ℃ 40 s,60 ℃ 30 s,72 ℃ 2 min,30个循环;72 ℃ 5 min。PCR产物通过1%琼脂糖凝胶电泳检测。序列测序并进行相似性比较(http://sw.ezbiocloud.net/ezeditor/)。利用ClustalX 1.81软件进行多序列比对,用MEGA 5.0软件计算进化距离,Neighbor-Joining法构建系统发育树。
1.4 菌株生长曲线与产酶性质测定 1.4.1 液体培养:将菌株接种到含5 mL产酶液体培养基的试管中,30 ℃、150 r/min培养约16 h。按1%的接种量转接到含有100 mL产酶培养基的三角瓶中。30 ℃、150 r/min摇床培养,培养至菌浓度不再变化,期间每隔12 h取菌液,测OD600吸光值并绘制生长曲线。 1.4.2 菌株木聚糖酶的测定:(1) 酶液准备:用1.4.1中的培养菌液,每隔12 h取1 mL菌液,经4 ℃、12 000 r/min离心10 min,收集上清液和菌体,菌体经裂解后8 000 r/min离心10 min取上清作为菌体粗酶液。(2) 酶反应体系:1%的燕麦木聚糖200 μL与50 μL稀释过的酶液充分混合;然后37 ℃恒温反应30 min;反应结束后,加入100 µL DNS溶液,在沸水浴中反应5 min后,立即放置于冰水混合物中;充分冷却后,补加蒸馏水650 μL使反应终体系为1 mL,测定OD520吸光值,做3个平行反应。同时做酶反应对照,在加入200 μL底物的EP管中先加100 μL DNS溶液,然后加入50 μL酶液,沸水浴反应5 min,之后步骤同前。(4) 酶活性定义:一个酶活性单位(U)定义为在37 ℃、pH 5.5的反应条件中,每分钟产生1 μmol还原糖所需要的酶量。 2 结果与分析 2.1 白蚁后肠木聚糖降解微生物的富集和分离纯化如1.2所述,通过选择培养基富集培养黄翅大白蚁后肠样品,104稀释度下的第2次富集培养液,在选择培养平板上长出约200个菌落。根据木聚糖降解圈大小,并结合刚果红染色筛选得到一个活性高的菌落,多次划线后获得纯菌株。在选择培养基上生长5 d的菌落,可直接看到降解圈(图 1A),刚果红染色结果如图 1B所示,纯化的菌株被命名为Mb1。
 |
| 图 1 培养于选择培养基的菌株Mb1 (A)和其刚果红染色(B) Figure 1 The clone Mb1 growing in the selective medium (A) and its congo red staining (B) |
| |
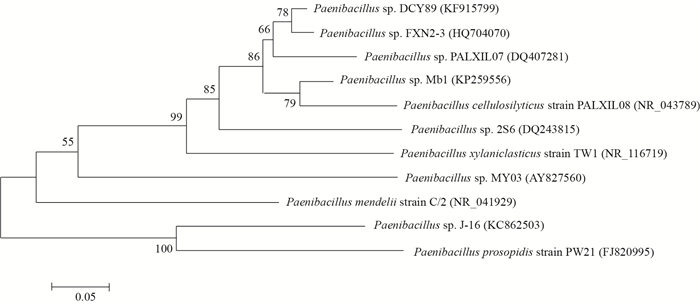 |
| 图 2 基于16S rRNA基因序列建立的Mb1和相关菌株的系统发育树 Figure 2 Phylogenetic tree based on the partial 16S rRNA gene sequence of Mb1 and other strains |
| |
对菌株Mb1在产酶培养基中的生长情况进行测定,从菌株Mb1的生长与产酶关系曲线可看出(图 3),培养24 h后细菌菌体浓度达到较高水平,72 h时菌体浓度最高,OD600吸光值为1.536,96 h时菌体浓度降至OD600值为1.166,此时胞外上清酶活为4.635 U/mL,在随后的培养中,菌体浓度维持在OD600值1.300左右,胞外上清酶活3.75 U/mL左右。细菌菌体浓度和胞外木聚糖酶酶活,在长达11 d的培养中基本保持稳定;另外同时所测的胞内菌体酶活保持在0.544 U/mL以下,提示菌株Mb1产生的木聚糖酶属于分泌型表达。Mb1在产酶培养基中生长时,前48 h由于培养基中含有蛋白胨,细菌快速生长,72 h时达到最大菌体浓度。在此过程中,上清中木聚糖酶活力处于较低水平,木聚糖利用不充分。随着Mb1继续培养,蛋白胨逐渐消耗殆尽,木聚糖作为培养基中的碳源,诱导菌体的木聚糖酶产生增加,并在随后细菌的培养过程中保持酶活稳定(图 3)。
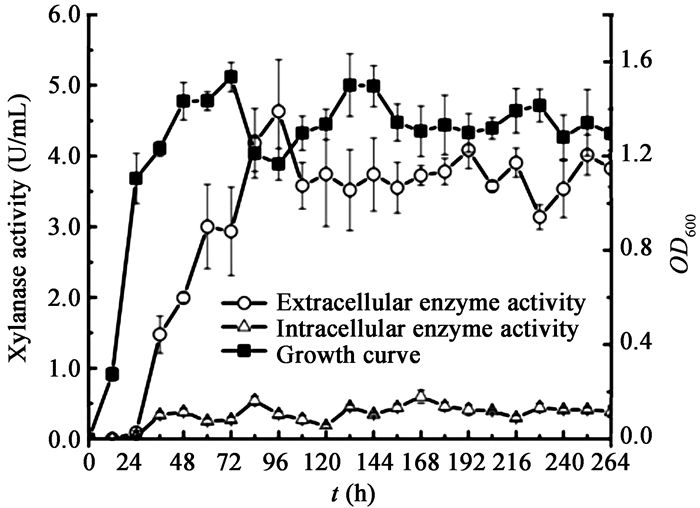 |
| 图 3 Mb1生长及木聚糖活性曲线 Figure 3 Growth curve and xylanase activity of Mb1 |
| |
本文从高等培菌白蚁——黄翅大白蚁后肠分离到一株具有较强木聚糖降解能力的类芽孢杆菌(Paenibacillus sp. Mb1)。类芽孢杆菌属于厚壁菌门,芽孢杆菌纲,芽孢杆菌目,芽孢杆菌科,类芽孢杆菌属。454高通量测序表明厚壁菌门是培菌白蚁后肠优势微生物之一,但目前为止尚没有该门微生物从培菌白蚁直接分离的报道[17]。已知具有木聚糖降活性的类芽孢杆菌可以从污泥、堆肥、纸浆黑液、森林土壤、稻田等多种环境中分离得到[18, 19, 20, 21, 22, 23, 24],另外有报道从食木白蚁肠道中分离到一株类芽孢杆菌[25]。培菌白蚁是一类体外与真菌共生而体内与细菌共生的高等白蚁[26],培菌白蚁、真菌和细菌三者如何协同降解木质纤维素一直是个谜。早期研究报道真菌给培菌白蚁提供纤维素酶[27],最近研究发现培菌白蚁自身可分泌产生纤维素降解酶,如内切葡聚糖酶和β-葡萄糖苷酶[28, 29],但尚未有白蚁自身分泌木聚糖酶的报道,而近年来的宏基因组研究报道培菌白蚁后肠有丰富的木质纤维素降解酶基因[13, 30]。本研究通过活性筛选的方法,直接从黄翅大白蚁肠道分离到产生木聚糖活性的类芽孢杆菌,暗示该微生物分泌的木聚糖酶可能参与黄翅大白蚁后肠的半纤维素降解过程,其作用可能类同低等白蚁后肠原生动物产生的木聚糖酶[9]。对类芽孢杆菌Mb1的进一步研究有助于了解培菌白蚁木质纤维素降解机制。
类芽孢杆菌Mb1是从以桦木木聚糖(Birchwood xylan)为唯一碳源的选择平板上筛选出来的,但是Mb1对多种木聚糖底物包括燕麦木聚糖(Oatspelt xylan)、榉木木聚糖(Beechwood xylan)都有降解活性。类芽孢杆菌Mb1分泌产生胞外木聚糖酶,与之前报道的白蚁肠道木聚糖降解菌相比,木聚糖酶活保持较长时间[25]。目前报道的木聚糖酶活是在基本培养基条件下获得的酶活,通过调整培养基中碳源和氮源比例等方法优化培养基,并将培养条件最适化,酶活有望进一步提高。
| [1] | Polizeli MLTM, Rizzatti ACS, Monti R, et al. Xylanases from fungi: properties and industrial applications[J]. Applied Microbiology and Biotechnology, 2005, 67(5): 577-591 |
| [2] | Prade RA. Xylanases: from biology to biotechnology[J]. Biotechnology and Genetic Engineering Reviews, 1996, 13(1): 101-132 |
| [3] | Ahmed S, Riaz S, Jamil A. Molecular cloning of fungal xylanases: an overview[J]. Applied Microbiologyand Biotechnology, 2009, 84(1): 19-35 |
| [4] | Deesukon W, Nishimura Y, Sakamoto T, et al. Purification, characterization of GH11 endo-β-1, 4-xylanase from thermotolerant Streptomyces sp. SWU10 and overexpression in Pichia pastoris KM71H[J]. Molecular Biotechnology, 2013, 54(1): 37-46 |
| [5] | Oliveira JV, Borges TA, Corrêa Dos Santos RA, et al. Pseudozyma brasiliensis sp. nov., a xylanolytic, ustilaginomycetous yeast species isolated from an insect pest of sugarcane roots[J]. International Journal of Systematic and Evolutionary Microbiology, 2014, 64(Pt 6): 2159-2168 |
| [6] | Hwang IT, Lim HK, Song HY, et al. Cloning and characterization of a xylanase, KRICT PX1 from the strain Paenibacillus sp. HPL-001[J]. Biotechnology Advances, 2010, 28(5): 594-601 |
| [7] | Rashamuse KJ, Visser DF, Hennessy F, et al. Characterisation of two bifunctional cellulase-xylanase enzymes isolated from a bovine rumen metagenome library[J]. Current Microbiology, 2013, 66(2): 145-151 |
| [8] | Yamada A, Inoue T, Wiwatwitaya D, et al. Carbon mineralization by termites in tropical forests, with emphasis on fungus combs[J]. Ecological Research, 2005, 20(4): 453-460 |
| [9] | Arakawa G, Watanabe H, Yamasaki H, et al. Purification and molecular cloning of xylanases from the wood-feeding termite, Coptotermes formosanus shiraki[J]. Bioscience, Biotechnology, and Biochemistry, 2009, 73(3): 710-718 |
| [10] | Tarayre C, Brognaux A, Bauwens J, et al. Isolation of amylolytic, xylanolytic, and cellulolytic microorganisms extracted from the gut of the termite Reticulitermes santonensis by means of a micro-aerobic atmosphere[J]. World Journal of Microbiology and Biotechnology, 2014, 30(5): 1655-1660 |
| [11] | Warnecke F, Luginbühl P, Ivanova N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite[J]. Nature, 2007, 450(7169): 560-565 |
| [12] | Nimchua T, Thongaram T, Uengwetwanit T, et al. Metagenomic analysis of novel lignocellulose-degrading enzymes from higher termite guts inhabiting microbes[J]. Journal of Microbiology and Biotechnology, 2012, 22(4): 462-469 |
| [13] | Liu N, Yan X, Zhang ML, et al. Microbiome of fungus-growing termites: a new reservoir for lignocellulase genes[J]. Applied and Environmental Microbiology, 2011, 77(1): 48-56 |
| [14] | Stevenson BS, Eichorst SA, Wertz JT, et al. New strategies for cultivation and detection of previously uncultured microbes[J]. Applied and Environmental Microbiology, 2004, 70(8): 4748-4755 |
| [15] | Wu Y, Hou XF, Chi SL, et al. Isolation and characterization of a Bacillus cereus strain with cellulose-degradation activity from a termite-inhabiting soil[J]. Microbiology China, 2012, 39(5): 638-644 (in Chinese) 吴燕, 侯信锋, 迟绍丽, 等. 白蚁栖息环境中蜡状芽孢杆菌的分离及其纤维素酶活性分析[J]. 微生物学通报, 2012, 39(5): 638-644 |
| [16] | Yang G. Microbiology Experiment Course[M]. Beijing: Science Press, 2005 (in Chinese) 杨革. 微生物学实验教程[M]. 北京: 科学出版社, 2005 |
| [17] | Otani S, Mikaelyan A, Nobre T, et al. Identifying the core microbial community in the gut of fungus-growing termites[J]. Molecular Ecology, 2014, 23(18): 4631-4644 |
| [18] | Tachaapaikoon C, Tanasupawat S, Pason P, et al. Paenibacillus xylaniclasticus sp. nov., a xylanolytic-cellulolytic bacterium isolated from sludge in an anaerobic digester[J]. Journal of Microbiology, 2012, 50(3): 394-400 |
| [19] | Khianngam S, Tanasupawat S, Lee JS, et al. Paenibacillus siamensis sp. nov., Paenibacillus septentrionalis sp. nov. and Paenibacillus montaniterrae sp. nov., xylanase-producing bacteria from Thai soils[J]. International Journal of Systematic and Evolutionary Microbiology, 2009, 59(1): 130-134 |
| [20] | Lee JC, Yoon KH. Paenibacillus woosongensis sp. nov., a xylanolytic bacterium isolated from forest soil[J]. International Journal of Systematicand Evolutionary Microbiology, 2008, 58(3): 612-616 |
| [21] | Sánchez MM, Fritze D, Blanco A, et al. Paenibacillus barcinonensis sp. nov., a xylanase-producing bacterium isolated from a rice field in the Ebro River delta[J]. International Journal of Systematic and Evolutionary Microbiology, 2005, 55(2): 935-939 |
| [22] | Arora A, Krishna P, Malik V, et al. Alkalistable xylanase production by alkalitolerant Paenibacillus montaniterrae RMV1 isolated from red mud[J]. Journal of Basic Microbiology, 2014, 54(10): 1023-1029 |
| [23] | Fathallh Eida M, Nagaoka T, Wasaki J, et al. Isolation and characterization of cellulose-decomposing bacteria inhabiting sawdust and coffee residue composts[J]. Microbes and Environments, 2012, 27(3): 226-233 |
| [24] | Ko CH, Chen WL, Tsai CH, et al. Paenibacillus campinasensis BL11: a wood material-utilizing bacterial strain isolated from black liquor[J]. Bioresource Technology, 2007, 98(14): 2727-2733 |
| [25] | Dheeran P, Nandhagopal N, Kumar S, et al. A novel thermostable xylanase of Paenibacillus macerans ⅡPSP3 isolated from the termite gut[J]. Journal of Industrial Microbiology & Biotechnology, 2012, 39(6): 851-860 |
| [26] | Aanen DK, Eggleton P, Rouland-Lefèvre C, et al. The evolution of fungus-growing termites and their mutualistic fungal symbionts[J]. Proceedings of the National Academy of Sciences of the United States of America, 2002, 99(23): 14887-14892 |
| [27] | Martin MM, Martin JS. Cellulose digestion in the midgut of the fungus-growing termite Macrotermes natalensis: the role of acquired digestive enzymes[J]. Science, 1978, 199(4336): 1453-1455 |
| [28] | Wu Y, Chi SL, Yun C, et al. Molecular cloning and characterization of an endogenous digestive β-glucosidase from the midgut of the fungus-growing termite Macrotermes barneyi[J]. Insect Molecular Biology, 2012, 21(6): 604-614 |
| [29] | Ni JF, Wu Y, Yun C, et al. cDNA cloning and heterologous expression of an endo-β-1, 4-glucanase from the fungus-growing termite Macrotermes barneyi[J]. Archives of Insect Biochemistry and Physiology, 2014, 86(3): 151-164 |
| [30] | Zhang ML, Liu N, Qian CL, et al. Phylogenetic and functional analysis of gut microbiota of a fungus-growing higher termite: bacteroidetes from higher termites are a rich source of β-glucosidase genes[J]. Microbial Ecology, 2014, 68(2): 416-425 |
 2016, Vol. 43
2016, Vol. 43